Challenges in a Bioinformatics Open-Source Era
Over the last few years open-source has been increasingly becoming the norm, even in Bioinformatics. The number of high-quality applications which are freely available on GitHub and other Git providers is increasing, such as the pipelines that the Broad institute uses for production.
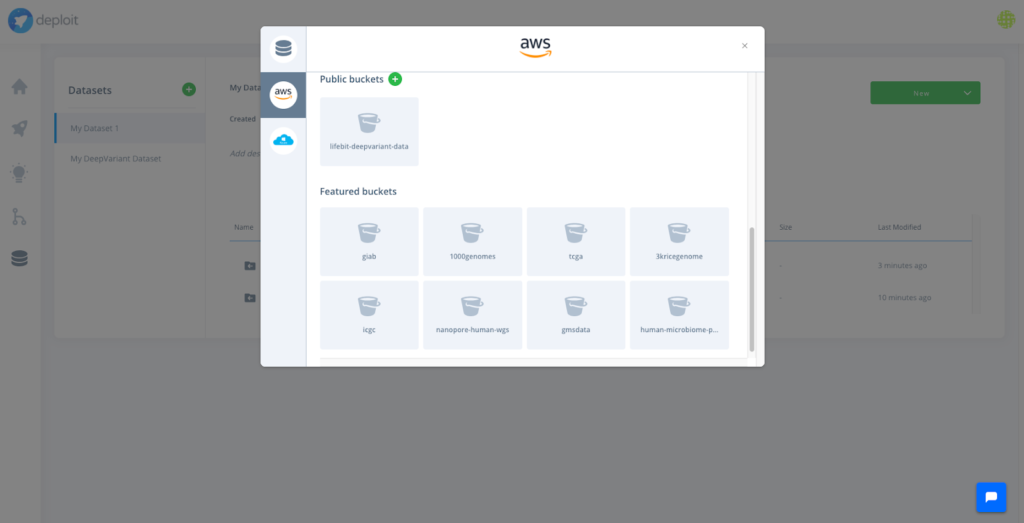
On the other hand, there has been considerable effort to collect publicly available datasets, across many different fields in biology/bioinformatics. The list is long but you can find one here of some databases containing data related to bioinformatics. Finally, some cloud providers are also putting at disposal biological datasets of interest, like AWS does with the Open Data project.
All of this is thrilling – such great resources becoming freely available gives room to a lot of new research and applications!
Although the potential is there, the real revolution will come when all of this can be brought together and produce insights that drive us forward.
The challenge is that bringing it all together, most of the time, is not easy. Data lives on different cloud storage providers (AWS S3 storage, Azure blob storage etc.), public available databases or on-premise storage systems. Pipelines can be found on different code versioning platforms (GitHub, Bitbucket, among others) or different container hubs (DockerHub, Quay, among others). Lastly, various on-premise high performance systems or cloud providers are used for computational power.
So when we actually run analyses, we first have to collect everything we need and handle compatibility, accessibility and quality assurance of the collected resources before we can begin the analyses – which is the core part of our work.
CloudOS as the Connection Centre
 In the currently growing bioinformatics open-source era, CloudOS brings together data, applications and resources, which would otherwise be dispersed. CloudOS synchronises with all the major software development tools and cloud providers. This enables easy access and running of open-source pipelines on publicly available data on the cloud, no matter where the pipeline or data are stored or which cloud provider you use for computation. All of it has been packaged in a simple and frictionless user experience and interface that also enables people with minimal to zero computational training to run and scale analyses over cloud.
In the currently growing bioinformatics open-source era, CloudOS brings together data, applications and resources, which would otherwise be dispersed. CloudOS synchronises with all the major software development tools and cloud providers. This enables easy access and running of open-source pipelines on publicly available data on the cloud, no matter where the pipeline or data are stored or which cloud provider you use for computation. All of it has been packaged in a simple and frictionless user experience and interface that also enables people with minimal to zero computational training to run and scale analyses over cloud.
At the moment, we are testing and making available a collection of high-quality pipelines that are popular among the community, off-the-shelf on the platform. We have already added a large list of popular Nextflow and Docker pipelines, and we will keep expanding it with more pipelines (Snakemake and CWL pipelines coming soon).
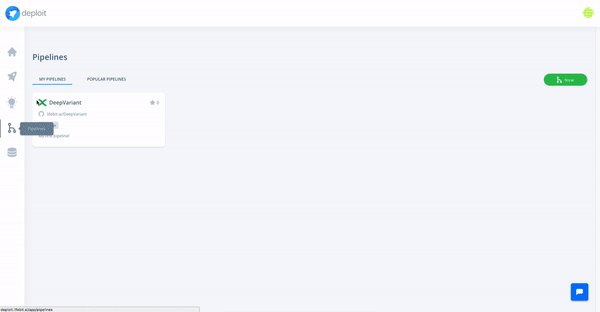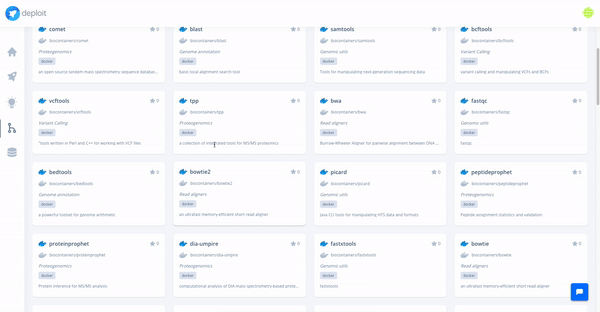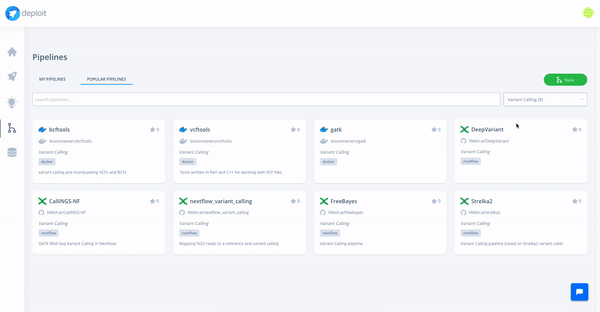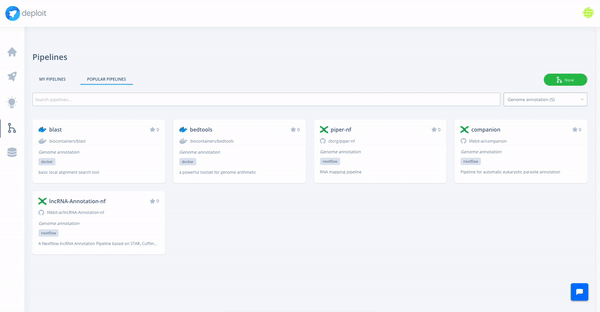
Moreover, we support import and visualisation of data from major cloud providers. Computation can be triggered through CloudOS on different cloud providers as well.
A Variant Calling use case
Variant Calling is a widely used tool, which identifies variants from sequence data. In this use-case it will be shown how to run FreeBayes as a Nextflow pipeline (an open-source variant caller) on publicly available data over cloud with CloudOS.
Open-Source Pipeline: FreeBayes
For this example, we are going to use FreeBayes as a pipeline. In CloudOS, we also provide many other variant callers like DeepVariant and Strelka2 as Nextflow pipelines, as well as easy access to many other open-source pipelines.
Public Dataset: Genome in a Bottle!
 Although the quality of the developed methods is advancing, different variant callers use different algorithms so may still disagree on their predictions. This is when benchmarking becomes essential to assess the quality of the predictions produced in order to identify those that are meaningful for clinical practice. The Genome in a Bottle Consortium has released a set of high-confidence, experimentally validated set of variants publicly available in a S3 bucket. This public reference material is extremely precious and various studies and benchmarks have already been conducted based on these datasets (e.g. 1,2,3).
Although the quality of the developed methods is advancing, different variant callers use different algorithms so may still disagree on their predictions. This is when benchmarking becomes essential to assess the quality of the predictions produced in order to identify those that are meaningful for clinical practice. The Genome in a Bottle Consortium has released a set of high-confidence, experimentally validated set of variants publicly available in a S3 bucket. This public reference material is extremely precious and various studies and benchmarks have already been conducted based on these datasets (e.g. 1,2,3).
Bringing it all together with CloudOS
One of CloudOS’ strengths is the synchronisation and integration capability. In fact, it synchronises with the main cloud providers as well as software development tools.
In practical terms, this translates into a seamless and effortless user experience, enabling the user to create as many collections of datasets and pipelines as they wish in virtually zero time and cost.
Porting a public or private pipeline in CloudOS becomes as easy as providing and URL (copy-pasting) from GitHub, BitBucket or DockerHub.
Accessing desired data can be achieved by either a simple upload of the data or a synchronisation with private or public cloud storage. CloudOS employs an intelligent virtual filesystem that allows the data to be linked but never moved or copied to the user’s cloud storage. It also prevents duplications of the data that already live within the user’s cloud storage. This minimises cost and data accessibility and management time.
Through a synchronisation to your cloud account, CloudOS manages the computation and delivers to the end user the final results.
Let’s have a concrete look on how it works through an example: running an open-source variant caller on an open source dataset over cloud in a scalable and reproducible manner.
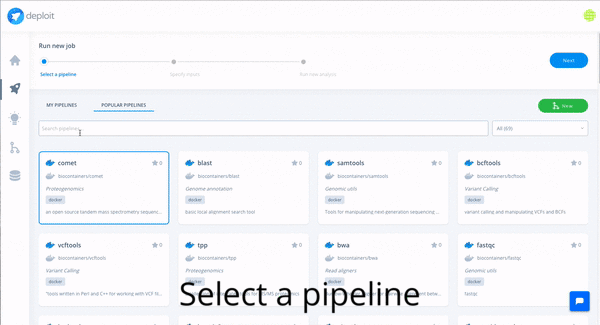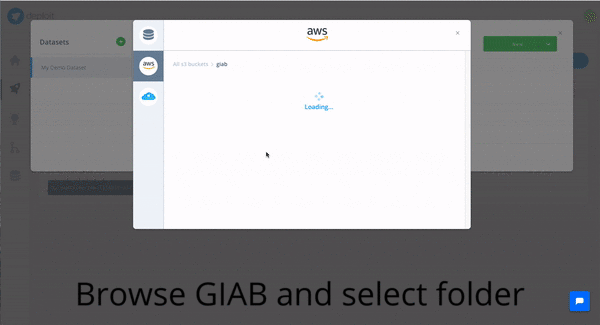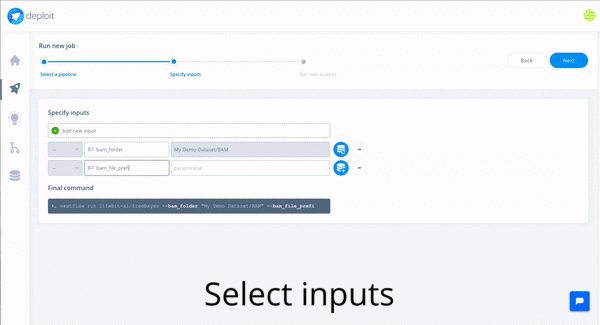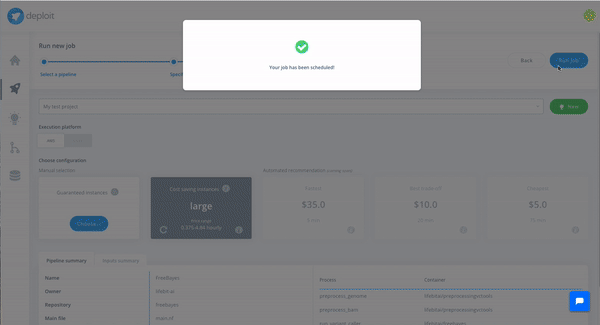
In order to do so with CloudOS, one of the featured variant callers can be selected: e.g FreeBayes. Afterwards, the content of the GIAB S3 bucket , as well an any other public S3 bucket, can be browsed, and bam files, the input required by FreeBayes, can be selected. Eventually, the computation is triggered by selecting the resource configuration. Done!
In the following gif you can observe how the whole journey looks like in CloudOS.
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!