Over the past few months, Lifebit has had an unparalleled opportunity to experiment and validate our ideas over one of the most innovative projects.
In Summer 2018 Innovate UK, part of UK Research and Innovation, announced the Design Foundation funding competition, investing up to £1 million to fund early-stage, human-centred research and design projects. The aim of this competition was to help businesses that wanted to explore opportunities to innovate based on the needs and behaviours of customers, users and stakeholders.
Lifebit joined forces with Science Practice, a life science design consultancy, where we came up with a project aimed at using human-centred design to understand market feasibility and user needs for an early-stage clinical gut microbiome AI-based analysis platform.
When results came in November 2018, we were delighted to find out that the Lifebit & Science Practice alliance got the grant!
Our team and I were thrilled to start the work on the project along with brilliant Science Practice designers and researchers Simon Hazelwood-Smith, Mar ek Kultys and James King, who brought their expertise in human-centred UX design and research specialised in the life sciences.
The Challenge
We were interested in launching a new gut microbiome analysis product on our platform but were unsure about the target audience for this type of solution. Our aim was to de-risk costly new product development by understanding existing market gaps and users needs in the gut microbiome space.
How we started
To shape our process we used the double-diamond framework: it allowed us to better understand customers and their pain points, while also exploring creative and innovative ways to solve their problems and delight them!.png?width=868&height=544&name=double-diamond%20(1).png)
Double Diamond consists of two approaches to problems and solutions: divergent and convergent thinking.
- Divergent thinking — thinking broadly, keeping an open mind, considering anything and everything.
- Convergent thinking — thinking narrowly, bringing back focus and identify one or two key problems and solutions.
Discover
This first step is all about divergent thinking, where we consider everything about a customer and their problem without constraints, challenging every part of the question and evaluating fields of interests.
It is a fact that our team knows a lot about the challenge, however that knowledge is naturally distributed. In the normal course of a project, having the chance to join forces and use all of that knowledge does not happen very often, but in this case, we made it happen.
To get started, we brainstormed the existing problems we could think of and generated answers in the form of HMW (How Might We) to solve these problems and dot-voted for the most important topics.


For secondary (desk) research where we focused on 3 main areas:
- Clinical pathways for microbiology testing in the NHS
- Consumer products for gut microbiome analysis
- Software for metagenomic analysis and visualisation
It was surprising to find out how many companies were already looking at these areas and trying to come up with solutions. Despite this, the market is still relatively new and unexplored.
In addition to secondary research, we also applied primary (field) research as well. The purpose of this round of primary research was to gather opinions on the following questions:
- What are the questions that need to be answered in clinical microbiology?
- What value could metagenomics provide to end users (clinicians in the healthcare context, consumers in the commercial context)?
- What are the possible routes of adoption of metagenomics in clinical practice?
- How does the Lifebit model fit into identified needs?
To do this, we created two sets of visual stimuli and a research protocol to go alongside the questions we were going to ask. Using both real-scenario models and assumption-scenario models, we aimed to trigger as many questions and thoughts as possible in a given timeframe.
.png?width=565&height=228&name=Stimuli-set-1%20(1).png)
.png?width=516&height=327&name=Stimuli-set-2%20(1).png)
We aimed to find 6-8 participants with both clinical and research backgrounds (50/50). For clinical experts, we looked amongst practising medical doctors working in infectious disease or microbiology departments at hospitals. For research experts, on the other hand, we looked amongst those who have expertise working in the healthcare context (i.e. epidemiology, population health), microbiome research and/or clinical trials. Luckily enough, we also had the chance to interview a commercial expert involved in the commercialisation of genomics testing services.
| #ClinicalMicrobiology #InfectiousDisease #MicrobialLab #MedicalMicrobiology | #DrugResistance #AntimicrobialResistance #Virology |
Method
We used semi-structured stimulus-based one-to-one remote or onsite interview research with the experts. There were two types of interviews depending on the individual’s availability:
While running the interviews, we were relentlessly taking recordings and interview notes:

Every interview was led by a facilitator, who was directing the discussion, deciding which questions were to be asked and showing the prototypes. The facilitator was always assisted by one or two notetakers who were managing interview recordings, taking notes and observing reactions.
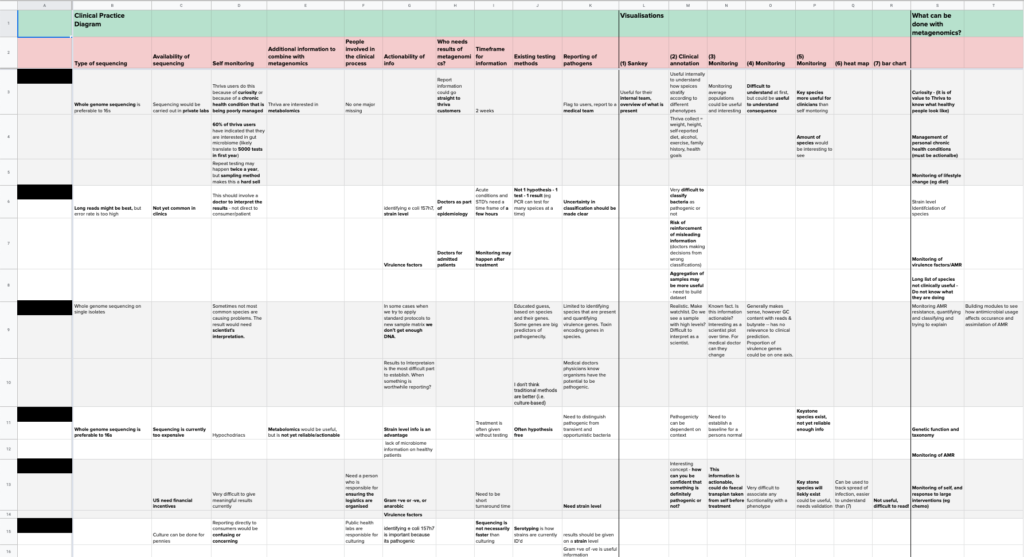
These golden dust materials subsequently turned into Table of Research Findings which became our holy grail and the basis to define the next steps.
Define
This step is to define the area of focus. Here, it is important to cluster learnings and similarities between themes to build opportunity areas. The result of DEFINE is to deliver a revamped brief, that either clarifies or contradicts the details.
We used our table of findings to do exactly that — generated Research Insights and decided on the direction for Develop.
Overall, the interviewees responded positively to the concept of using metagenomics in connection with clinical/wellbeing contexts. However, the majority expressed reservations about the maturity of the field, specifically to be able to make evidence-based, meaningful connections between metagenomic results and specific phenotypes.
We gathered again to get the most of the team’s expertise to come to the best possible decision. From the previous steps, we defined two potential ways of furthering research development:
| What? Self-monitoringResearch participantsPeople who are conscious of their health and are already monitoring, have performed other tests or are thinking about it. Research questionsWhat type of information is interesting to this type of user in the first place, and actionable in the long term. Design stimulusUI with options for connecting metagenomic data to phenotype informationOne-off report in monitoring service | What? Clinical researchResearch participantsResearchers / existing or new users / restricted to people working in antimicrobial resistance, for instance. Research questionsHow can we make it easier for clinical researchers to find out the clinical relevance of metagenomic data? How to start comparing populations? Design stimulusUI, graphs, research workflow |
While running interviews with metagenomics experts, we also found a strong interest from direct consumers to know more about how to improve their health, even though they did not have specific health-related problems. I dubbed this audience the “health tech innovators” – their goal is to bring efficiency to their energy levels to exceed the world’s current demands.
Finally, we decided to go with the self-monitoring option, as we considered this direction has the greatest potential.
Develop
With a vision in place, it was time to explore the best potential solutions. We knew what we wanted to achieve, and by exploring and validating options, we were trying to find the best ways to succeed.
As we had already deduced the actual question to solve or challenge, we started ideating.
This is the fun part, the diverging phase. We restrained from limiting ourselves and approached ideation with open minds. We let anything happen at this point and built upon each other’s ideas. After we combined the best ones, we carefully stapled them into one single paper prototype.
The paper prototype became the point of departure on the way to the more refined interface, which we later implemented on desktop and tablet and started testing with the users.
Prototype Gut Health Report
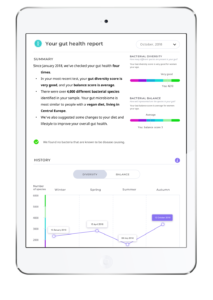
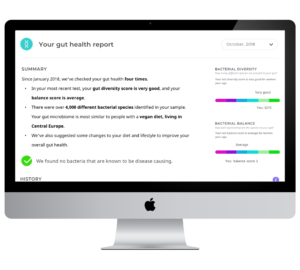
We were trying to keep a fair split between users with prior experience of quantitative health services and newbies – people who are motivated to use a personalised quantitative health service but have not yet done so. Besides potential users, we also got to talk to a professional in a product development role in a company providing personal quantitative health services, as well as another individual with expertise in the commercialisation of gut metagenomic testing.


As in the previous set of interviews, we produced the precious Table of Research Findings from the outcomes received.
%20(1)-1.png?width=868&height=494&name=7a0b9-table-of-findings-2%20(1)%20(1)-1.png)
Deliver
Deliver step is all about generating research insights from Develop and formulating design recommendations. In other words: providing conclusions from the project to inform further work.
In this phase, we’ve generated an Executive Summary.

It is a high-level overview of what users would expect from such a product, what would make it trustworthy, understandable and, most importantly, actionable.
The summary also contained design recommendations for each section, indicating the importance of MVP and current Design Maturity. Last but not least, it outlined the next steps for the product elaboration strategy.
Thanks to Marek and Simon, who are pros specialising in life science projects. I got invaluable experience and gained tons of knowledge while working on the project and learned some best practices. The application for the grant, planning, housekeeping and facilitating also involved: James King, Mike Furness, Claire-Alix Zapata, Pauline Richard, Phil Palmer, Pablo Prieto and Maria Chatzou.
My key takeaways were how to adjust the time spent on developing ideas and the time spent on prototyping, interview running techniques, as well as the process of converting raw interview data into an Executive Summary.
Stay tuned for more exciting developments! We have started an Early Access Program for companies and organisations interested in learning more about this brand new Clinical Gut Microbiome AI-based analysis platform. Find out more about the Early Access Program by filling out the following form .
_______________________________________________________
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!