In this two-part blog series, we delve into the challenges of secondary NGS analysis and bioinformatics and how Illumina’s DRAGEN and CloudOS come together to tackle these issues. Illumina’s DRAGEN, an accelerated and improved cloud-native (AWS) implementation of the standard BWA/GATK, resolves the issue of lengthy compute times and massive data volumes for the secondary NGS analysis of human genomes. In this second blog post, we give you a step-by-step play of how easy it is to run Illumina’s DRAGEN through CloudOS over your data, in your own AWS cloud.
CloudOS brings the power of Illumina’s DRAGEN to your data, in your environment.
Understanding the Importance of Secondary NGS Analysis
Secondary NGS analysis is crucial for understanding genomic data and its implications in precision medicine.
At Lifebit, we understand the importance of standardisation, reproducibility, auditability and making all stages of analysis FAIR. That is why we always prioritise adding features to the CloudOS platform that will enable users to access the best-in-class tools to standardise all aspects of their analysis. CloudOS makes Illumina’s DRAGEN easily accessible to anyone through the CloudOS Marketplace, at no extra cost, as users only need to cover the standard Illumina DRAGEN-AWS pay-as-you-go fees.
The advantages of running Illumina’s DRAGEN through CloudOS, instead of natively over AWS, are:
- Getting easy access to and deploying Illumina’s DRAGEN over your own AWS cloud
- Managing all data, environment and Illumina’s DRAGEN in one place
- Getting out-of-the-box versioning, 1-click cloning/reproducing and sharing of any analysis performed with Illumina’s DRAGEN
- Enabling real-time monitoring of the analysis progress, its owner, its resources utilisation, its cost, inputs and outputs
- Reducing the cost of running Illumina’s DRAGEN over AWS
- Enabling collaboration and better management of data analysis projects that involve Illumina’s DRAGEN alongside with other type of analysis
- Instead of moving your data to DRAGEN, you bring Illumina’s DRAGEN to your data.
When using CloudOS, you ultimately significantly decrease costs and turnaround times, while at the same time, vastly improve the accuracy, versioning/trackability and reproducibility of your secondary analyses.
How to setup Illumina’s DRAGEN in CloudOS
The integration of Illumina’s DRAGEN enhances the efficiency of secondary NGS analysis.
With secondary NGS analysis, researchers can derive significant insights from their genomic data.
Utilizing advanced techniques in secondary NGS analysis can lead to better variant understanding.
Here, we will walk you through the steps we took to run Illumina’s DRAGEN as a CloudOS module. In this case, the module takes care of running the current industry standard variant calling pipeline which follows BWA-MEM for read mapping and GATK HaplotypeCaller for variant calling in a sample. The input formats used for the pipeline are FASTQ files.
Pipeline cost breakdown
Since the CloudOS platform operates using a federated approach over your AWS cloud account, the costs related to the execution of the pipeline are solely explained by the usage of two elements:
By optimizing secondary NGS analysis, researchers can save time and resources effectively.
- EC2 Instance type cost
- Software license cost
Secondary NGS analysis plays a vital role in genomic research and clinical application.
Both elements are pay-as-you-go and are directly billed to you through the AWS billing system. For instance, if you run Illumina’s DRAGEN in a f1.16xlarge instance for a single 30x dataset for ~30 minutes of compute, the total cost would be around $18, with the following breakdown:
- $11 EC2 costs for f1.16xlarge
- $7 Illumina’s DRAGEN Software license
Getting started with Illumina’s DRAGEN in CloudOS
In order to run the Illumina DRAGEN pipeline, you only need to cover the following two requirements:
When considering secondary NGS analysis, it is essential to utilize reliable tools like Illumina’s DRAGEN.
- Having an AWS cloud account at hand to use to setup your account in CloudOS
- Subscribing to the Illumina DRAGEN software in your AWS Marketplace
1. Setting up access in CloudOS
Understanding the intricacies of secondary NGS analysis can greatly enhance research outcomes.
Before running any pipeline, you can follow the general guidelines on how to register and synchronise credentials from an AWS account into the CloudOS platform.
Secondary NGS analysis requires a robust framework to support complex genomic data interpretation.
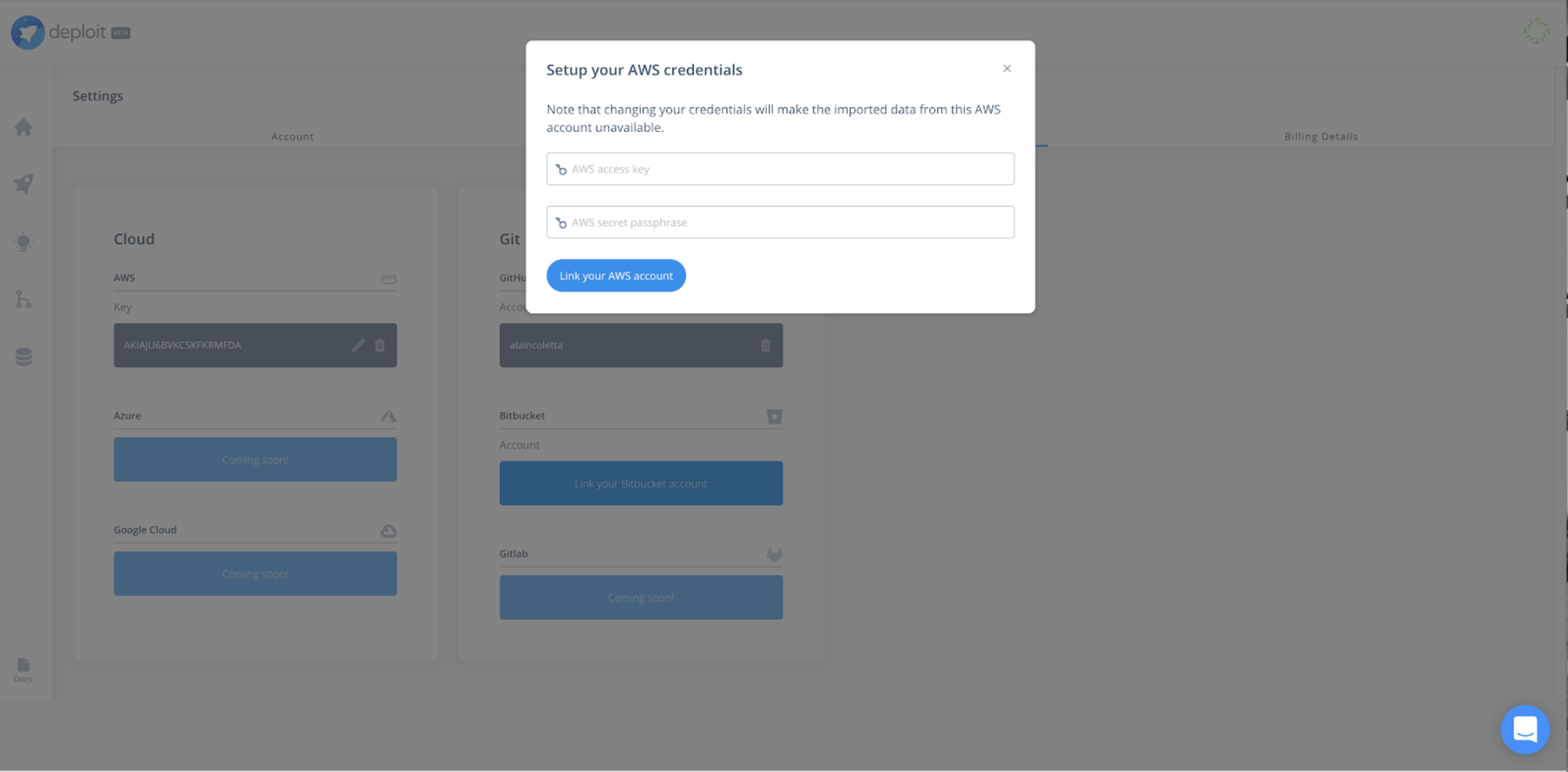
In order to connect an existing cloud to CloudOS, you simply add your AWS user credentials to provide permissions to manage cloud resources on your behalf and orchestrate Illumina DRAGEN analysis. For more information, and a step by step guide on how to correctly setup cloud credentials, refer to this documentation.
2. AWS Marketplace subscription
Implementing cloud solutions for secondary NGS analysis enhances accessibility and collaboration.
Before trying out Illumina’s DRAGEN, make sure you have correctly connected your AWS cloud to CloudOS. In order to start using Illumina’s DRAGEN, you have to subscribe to DRAGEN, which allows programmatic access to deploy Illumina’s DRAGEN within your user cloud resources. The Illumina DRAGEN software is accessible to any AWS user through the AWS Marketplace.

For more information on the usage of Illumina’s DRAGEN within AWS cloud resources, refer to the following guide.
With CloudOS, secondary NGS analysis becomes streamlined and efficient, allowing for rapid processing.
After successfully completing the two previous steps, you will be able to run a fully cloud-managed Illumina DRAGEN module in CloudOS from the Marketplace section. CloudOS takes care of managing the following aspects related to the deployment and execution of Illumina DRAGEN software:
Secondary NGS analysis simplifies the identification of variants and helps in comprehensive genomic understanding.
- Allocation of compute resources within your AWS cloud
- Setup of network & compute resources
- Scheduling of Illumina DRAGEN analysis
- Monitoring of analysis progress and costs
- Versioning of the analysis
- 1-click cloning and sharing of the analysis
- Federated data access
- Data management
The integration of Illumina’s DRAGEN and Lifebit CloudOS within your cloud environment makes it possible to automate and run federated data analysis with Illumina’s DRAGEN and CloudOS in only a few clicks. This makes it ideal to be used by data scientists that have no experience on how to deploy such analysis over cloud but also by experts that want to be optimising and automating such analysis deployment to save time and money.
Running Illumina’s DRAGEN in CloudOS
The benefits of secondary NGS analysis are amplified with the use of high-performance cloud computing.
CloudOS provides a Marketplace similar to the AWS Marketplace, accessible to any CloudOS user, and includes several Illumina DRAGEN pipeline modules. Each Illumina DRAGEN module in CloudOS comes with real-time monitoring and reports, which provide insights on alignment and variant calling metrics for each sample processed, including interactive charts.
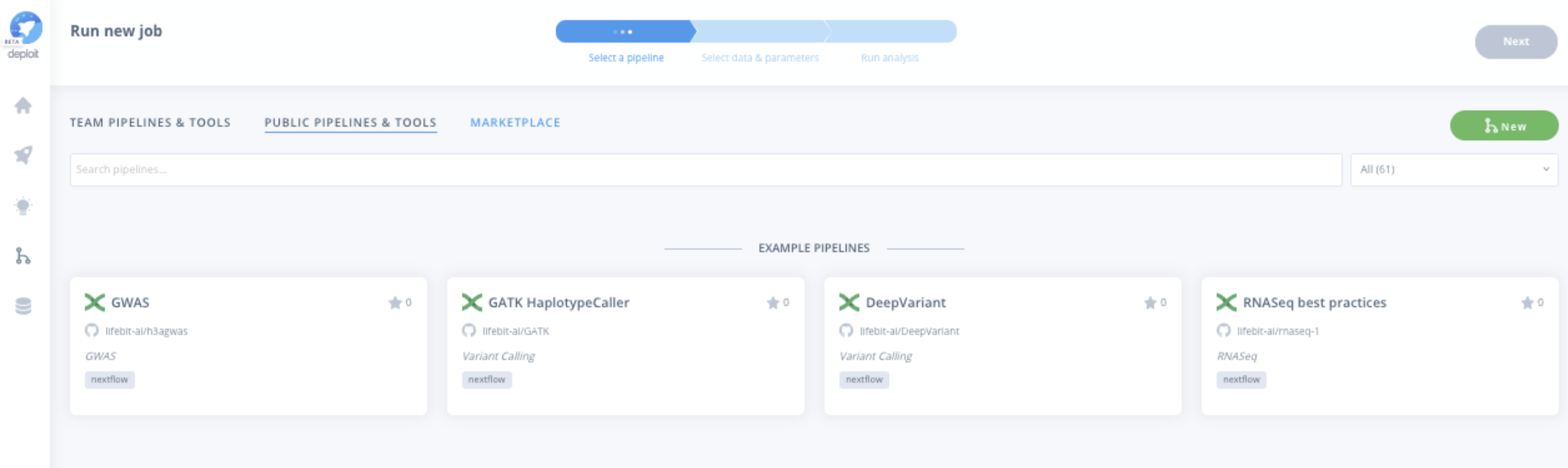

Secondary NGS analysis is essential in identifying clinically relevant mutations for patient care.
For the germline variant calling module, you will be prompted to provide the basic input options for running an analysis:
- RAW FASTQ files containing the read sequences for each sample, and
- the reference genome against which alignment and germline variant calling will be performed.
Collaborative efforts in secondary NGS analysis can lead to breakthroughs in personalized medicine.

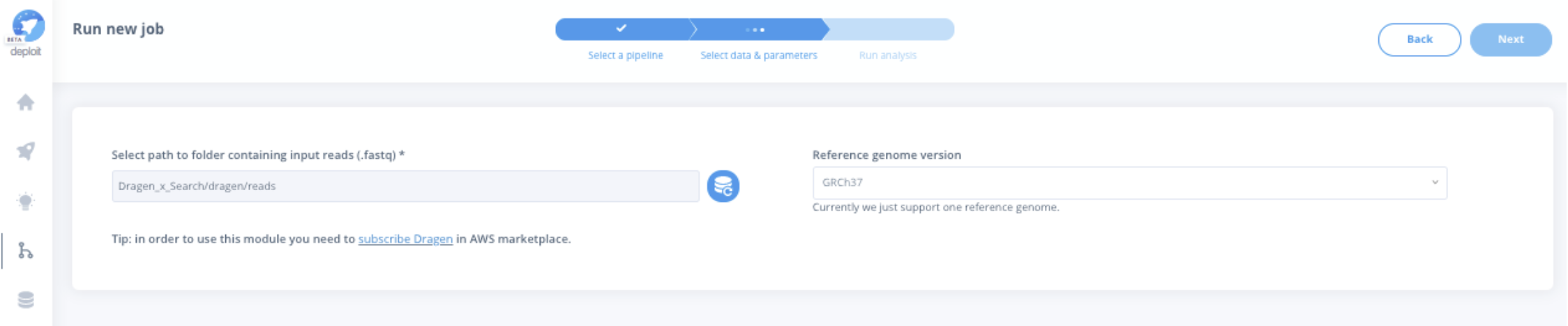
Through secondary NGS analysis, researchers can better understand genomic alterations and their impacts.
Once the analysis has been launched, CloudOS will track, in real-time, the execution and provide metrics on cloud consumption and cost estimates according to the resources being used in your AWS account and the Illumina DRAGEN software license you subscribed to in the AWS marketplace.

The ongoing advancement in secondary NGS analysis technology enhances data quality and reliability.
After an analysis is completed, the sample variant calls in VCF format, the metrics and logs of the Illumina DRAGEN analysis execution are stored and directly available in your cloud storage account. CloudOS always manages the data transfer within your cloud account, as CloudOS runs all analysis in a federated manner, bringing compute to data, never allowing the data to leave your cloud environment or control.

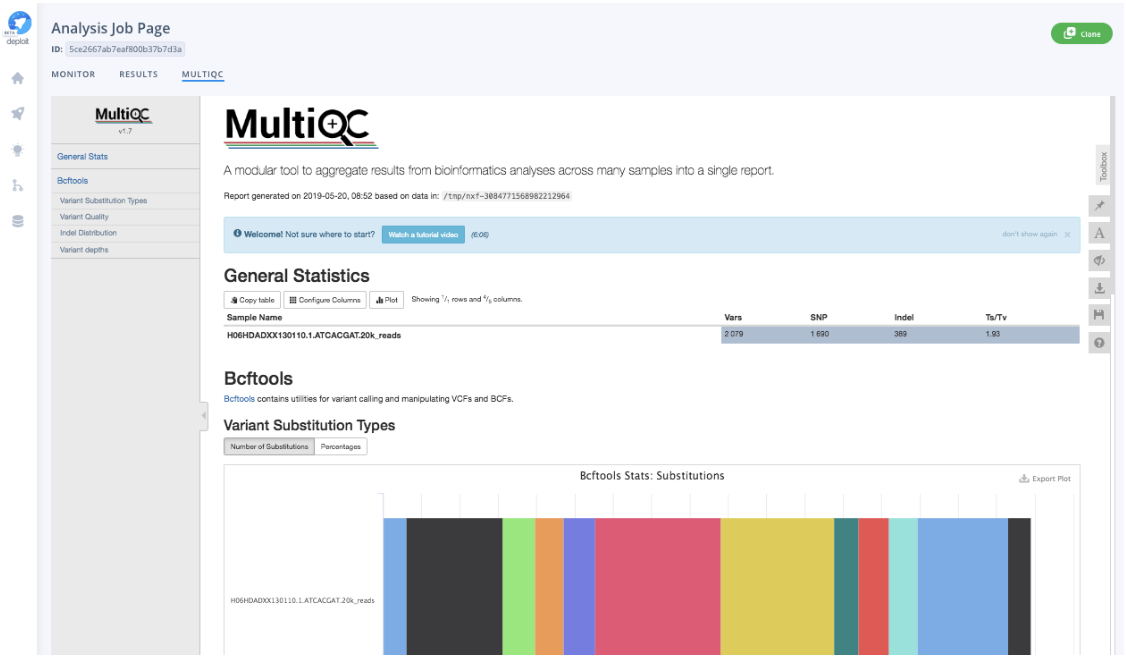
Additionally, once the analysis has been finalised, you can directly visualise a MultiQC report that summarises metrics of the sample variant calls by Illumina’s DRAGEN. The metrics in the report serve as a direct quality control (QC) step, and include variant call quality, distribution of variant types and depth across all samples.
Conclusions
As you can see from this step-by-step tutorial, running Illumina’s DRAGEN on CloudOS is straightforward and requires minimal setup on your part. By using CloudOS to run Illumina’s DRAGEN, you get end-to-end versioning and real-time monitoring of how your analysis is progressing, in addition to tracking which resources are used and the costs incurred during the run. This is a significant improvement on running Illumina’s DRAGEN natively through your AWS account, as this essential auditing is not available otherwise.
Furthermore, if you are part of a team and need to share results from your Illumina DRAGEN run, you can easily do so with the 1-click sharing button on the analysis job page. This effectively removes silos between researchers in the same team, or collaborators from different organisations. Moreover, if the run needs repeating, you can simply clone the analysis by using the 1-click cloning feature on the analysis job page, ensuring true reproducibility between runs.
Lastly, by running Illumina’s DRAGEN directly in your own environment in a federated manner, your data’s safety is guaranteed: instead of moving your data to Illumina’s DRAGEN, you bring DRAGEN to your data.
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!
As technology evolves, the methods of secondary NGS analysis continue to improve, providing better insights.
The future of secondary NGS analysis looks promising with the integration of AI and machine learning.