Key Features of a Trusted Research Environment

Key Features of a Trusted Research Environments
In the last twenty years, there has been an explosion in the production of patient-derived biomedical data. This includes datasets derived from clinical-genomic, Electronic Health Records (EHRs), and real-world data (RWD) sources, which, when utilised together, can hold the answers to the underlying causes of disease. Unfortunately, the transformative potential of this health data has yet to be realised. To preserve patient privacy, much of the world’s health data is stored within institutional siloed environments that are unavailable to researchers or are difficult to access. To support research and innovation through the power of data, solutions are needed to enable data access and linkage while maintaining security.
Trusted Research Environments are highly secure and controlled computing environments that solve this problem. Also known as “Data Safe Havens” or “Secure Data Environments”, trusted research environments allow approved researchers from authorised organisations a safe way to access, store, and analyse sensitive data remotely.
Across biobanking and health sectors, trusted research environments are becoming increasingly prevalent as a means to achieve both data accessibility and security. For example, the UK-national biobank Genomics England currently hosts data from over 135,000 NHS patients within a trusted research environment for approved research use. Further, the National Health Service (NHS) in the UK is actively investing in developing trusted research environments to power life-saving research – forming an open and innovative health data ecosystem that will improve productivity in uncovering disease treatments.
There are many advantages to using trusted research environments, and with their potential to progress research and benefit patients’ lives, it is exciting to see their implementation across the healthcare sector. However, as trusted research environments continue to be adopted, it is essential to ensure that they are developed with security by design, so that misuse of personal data is never a concern. This article presents the key features of a trusted research environments to both maximise research potential while ensuring data security.
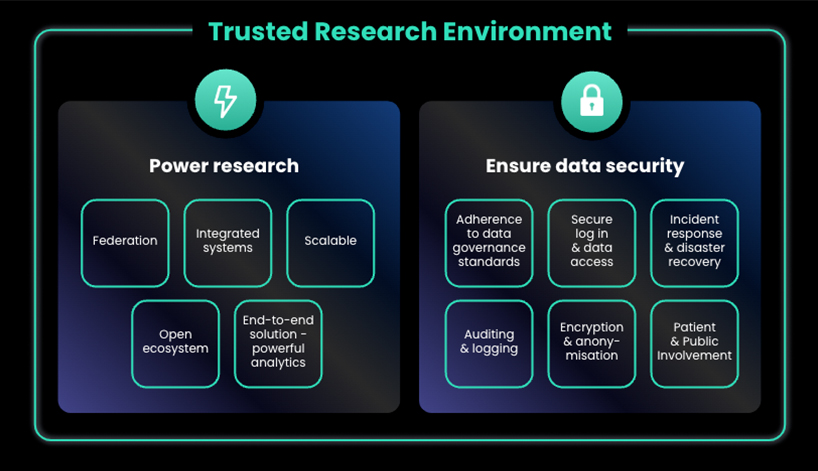
Key Features of a Trusted Research Environments to Power Research
To accommodate increasing volumes of data, trusted research environments must first be scalable. Biobanks with hundreds of thousands of datasets must quickly scale to house petabytes in volume. However, scalability can be a challenge when it comes to cost, or being able to quickly configure extra storage as datasets grow and highly performant compute is required. A solution to this is cloud-based trusted research environments; with the “elastic” nature of cloud computing, trusted research environment owners, such as biobanks, only pay for the storage they require, which is more economically sustainable.
A trusted research environment should further integrate with diverse sources and systems where the data is being generated. Automatic data ingestion from multiple sources will avoid backlogs in data transfers that can ultimately slow progress. A trusted research environment integrated into various data sources also enables researchers to seamlessly utilise multiple data types – for example, access to both genomic and phenotypic data can help scientists link molecular changes to disease presentation.
When integrating data from various sources, it is crucial to consider the risks associated with physically moving data. Not only can it become expensive and time-consuming, but moving or copying data into a centralised trusted research environment poses security risks – removing control from the data custodian and jeopardising patient privacy. Federation capabilities are an innovative, critical feature of trusted research environments that simplifies the linking of disparate data sources without physically having to move the data itself.
Within a federated architecture, data will remain within appropriate jurisdictional boundaries, while metadata are centralised and searchable. This is an alternative to the traditional model for data access, in which data is moved or duplicated to be centrally housed. Over the last five years, there has been a shift in individual organisations connecting to form federated data platforms, enabling data access and computation for users across organisations.
Another essential feature is an open ecosystem approach to trusted research environments and federated data platforms. Typically, developed trusted research environments can be classified as either Closed, Do-it-yourself (DIY), or open ecosystem:
- Closed trusted research environments are typically defined as proprietary software that is distributed under a licensing agreement where authorised users can only view inputs and outputs of their analyses – as a result, vendor-lock-in is common. Therefore, some organisations choose to build their own trusted research environment from scratch – a DIY approach.
- DIY can appeal to organisations looking to leverage the latest innovative open-source software and incorporate it into their research and development pipelines. However, organisations can become heavily reliant on the developer community for support (facing significant challenges around work continuity), and best-practice coding and testing standards may not be achieved.
- Open platforms provide the reliability and support of closed platforms, as these are usually distributed under a licensing agreement. Open platforms also offer end users the endless possibilities of a DIY platform in customising their environment by integrating with third-party applications and tools.
Finally, trusted research environments must not simply house data but should also incorporate analytics. The data within a trusted research environment must be ready to use for research purposes. It is far too common for researchers to spend an inordinate amount of time searching for and preparing their data before they can perform valuable analyses that could benefit patients. This is a requirement when data has come from disparate sources; for example, clinical data often incorporates varied medical vocabularies when collected at different institutions, which must be standardised before the datasets can be used in further analysis.
ETL (Extraction, Transfer, Loading) pipelines that are built into a trusted research environment can automatically standardise the ingested data to a common format, such as the internationally-utilised OMOP Common Data Model. FAIRifcation of data within the trusted research environment further makes data Findable, Accessible, Interoperable and Reusable by incorporating unique identifiers for data and metadata management. With automatically standardised data, researchers will save time and immediately be able to use the data within a trusted research environment for their research purposes.
Once the data is in a usable format, trusted research environments should incorporate built-in, reproducible analytical pipelines (RAPs) to support users in transforming analysis-ready data into insights in a robust manner. Genomics England’s trusted research environment includes integrated, open-source tools to enable researchers to analyse the data housed within the trusted research environment. Further, low/no-code tools should be integrated within trusted research environments to support all end-users, regardless of their coding proficiency or data science background. Between securely housing the data, standardising it to a usable format, and offering tools to derive valuable insights, trusted research environments can provide an end-to-end solution to enable rapid and efficient research progress.
Key Features of Trusted Research Environments to Ensure Data and Patient Security
With all that trusted research environments can offer to help progress research, there are also crucial features necessary to ensure data security. With the increasing adoption of trusted research environments worldwide, there are emerging data governance standards that outline how trusted research environments should be operated. At a UK-national level, the UK Health Data Research Alliance, convened by Health Data Research UK (HDR UK), has adopted a set of principles to ensure data services, like trusted research environment providers, provide safe research access to data. These are based upon the Five Safes Framework, initially established by the Office of National Statistics, and now broadly adopted across the international research community:
Key Features of a Trusted Research Environments for Enhanced Research Security
Safe data – Confidentiality of data is maintained
Safe projects – Data owners approve the research projects
Safe people – Researchers are trained to use the data safely
Safe settings – A secure computing environment prevents unauthorised data access
Safe outputs – All exported results are screened and approved
Below the national level, well-defined governance frameworks should outline the roles and responsibilities of different stakeholders, including researchers, institutional review boards, and information security teams, to ensure that patient data are handled responsibly. This can become increasingly complex, with data governance standards rapidly changing across regions and between institutions. Working with a reputable trusted research environment provider can alleviate these complications. When choosing a provider, certifications in industry-recognised standards, including ISO 27001 and Cyber Essentials Plus signify that the provider is equipped to manage private and sensitive data.
Within the trusted research environment itself, key security features will ensure only authorised individuals can access sensitive patient data. These processes can include using secure login procedures, such as two-factor authentication, and implementing role-based access controls, which limit access to data based on an individual’s role or responsibilities. Within the trusted research environment, all data stored should be encrypted so that data can only be accessed by those with the correct decryption key. When researchers have been given access to a dataset, all identifiable information should be pseudonymised/anonymised to maintain patient privacy. Together, these efforts help protect sensitive data from unauthorised access and privacy breaches.
Despite secure-login procedures, role-based access, data encryption, and anonymisation, there remains a concern regarding malicious or inappropriate use of patient-derived data among members of the public and privacy groups. Auditing capabilities are a critical feature of trusted research environments that can minimise these risks, as no action within the trusted research environment can be taken without being recorded. Routine auditing can review data access logs to identify unusual activity, such as attempts to access data without authorisation. Ongoing network traffic, user activity, and other actions can further be monitored to detect potential security incidents in real-time. Should an unauthorised incident or data breach occur, a trusted research environment should have a robust incident response and disaster recovery plans in place.
Finally, meaningful patient and public engagement in how trusted research environments should manage biomedical data are becoming an industry standard to ensure the risks of data misuse are minimised and that the data research is focused on public benefit. Many examples demonstrate how patient and public involvement in decision-making on appropriate data use and access through trusted research environments are leading to improved research output.
National initiatives to collect valuable patient biomedical data are essential to progress research, yet they must do so with public interest at the forefront. It is promising to see that National Initiatives themselves have agreed to recommend patient engagement frameworks to help govern how that data should be managed. For example, Genomics England conducts routine patient and public engagement to help shape their genomic medicine programmes. Promoting patient and public trust can help ensure the longevity of the programmes that trusted research environments support, helping progress research efforts long-term.
Trusted research environments are emerging as essential entities that can scale with increasing volumes of patient data and ensure its protection, all while enabling secure access for approved research. Lifebit is proud to implement the key features described above in our work with valued clients, including Genomics England, the Danish National Genome Centre, Boehringer Ingelheim, NIHR Cambridge Biomedical Research Centre, and others. Lifebit works proactively with clients to comply with sensitive data requirements. Lifebit ensures that organisations can meet and exceed industry standards amidst the changing regulatory and regional landscape – enabling valuable research at scale to improve patients’ lives.
To find out more:
Read The Complete Guide to Trusted Research Environments in 2023
Read Lifebit’s whitepaper on best practices for building a Trusted Research Environment
Read Lifebit’s whitepaper on security and data governance
Request a platform demo
Email us at hello@lifebit.ai

