The Data Symphony: Harmonizing Health Information for Global Impact

Stop Losing 6 Months and 30% of Your Data: Harmonize Health Data to Speed AI and Care
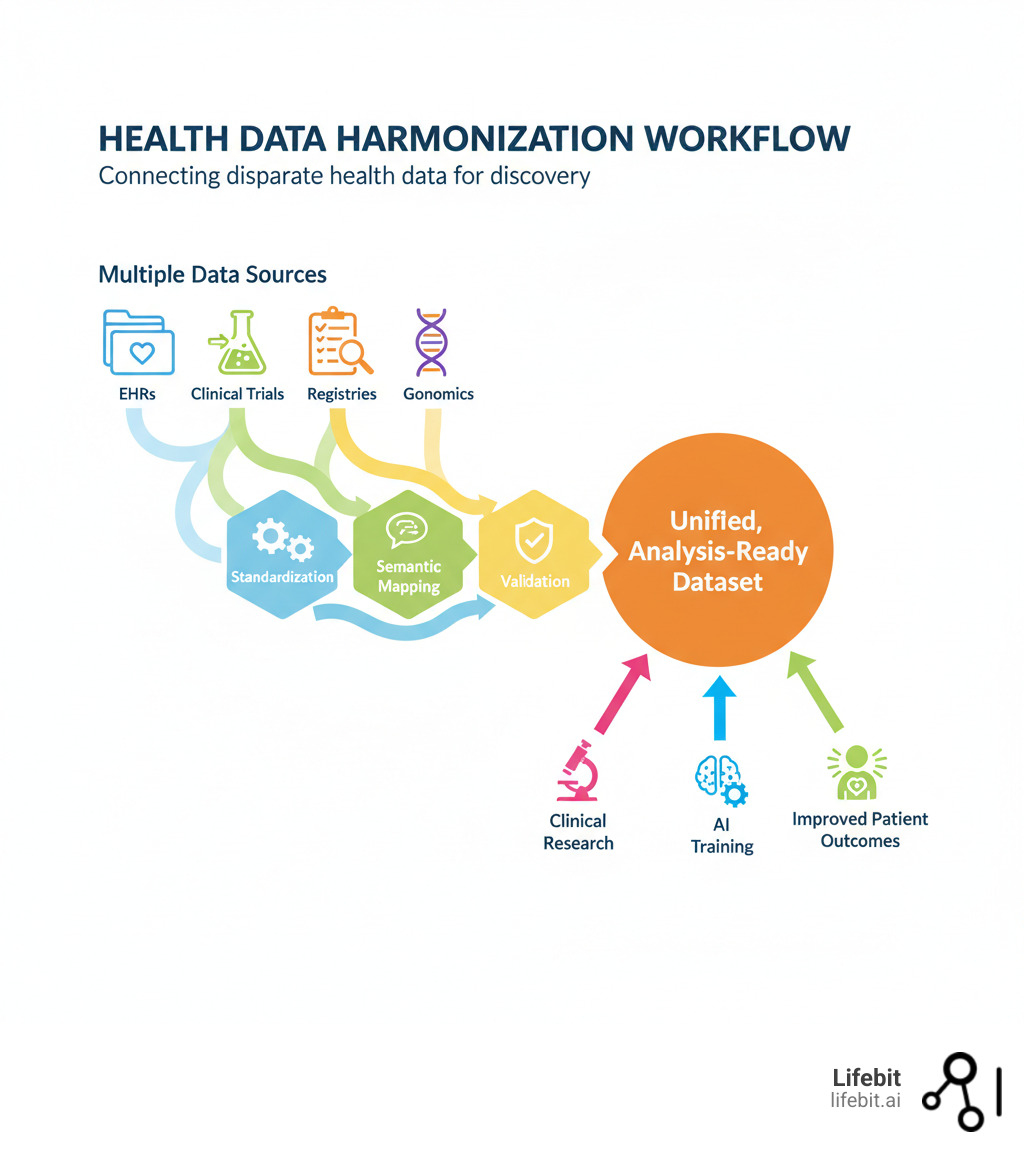
Health data harmonization is the process of making data from different sourceslike EHRs, clinical trials, and genomics databasescomparable and interoperable for analysis. It involves resolving differences in data formats (syntactics) and meaning (semantics), mapping between standards like HL7 FHIR and OMOP CDM, and preserving clinical context.
The digital change of healthcare created vast but disconnected data repositories. This creates a fundamental problem: AI models require massive datasets for training, but organizations collect data differently. Attempting to combine these datasets without proper harmonization leads to errors, bias, and wasted resources. Project-specific efforts can take up to six months, with nearly a third of the data often discarded because it can’t be reconciled.
The stakes are high. Without harmonization, research is delayed, resources are wasted on data wrangling, and grant funding is jeopardized. For rare disease research, where combining data from multiple organizations is essential, harmonization is critical for patient survival.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit. For over 15 years, my work in federated data analysis and computational biology has focused on solving these challenges to enable precision medicine through unified biomedical data.
Health data harmonization terms simplified:
Why Your Health Data is Trapped in Silos
Imagine trying to solve a jigsaw puzzle where every piece comes from a different box. That’s what healthcare organizations face when combining data. The promise of big data—from accelerating drug discovery to personalizing care—hits a wall: health data harmonization challenges that lock valuable information in isolated systems.

Several key issues make data integration so difficult:
- Syntactic differences: Data arrives in incompatible formats like CSV, JSON, or XML. One system might export patient records as a flat file, while another uses a complex, nested JSON structure via an API.
- Semantic ambiguity: The same term can mean different things. “Blood pressure” might refer to a systolic reading, a diastolic reading, or an average, with the context lost. A diagnosis of “diabetes” could mean Type 1, Type 2, or gestational diabetes, but if the specific code is missing, the data is unusable for targeted research. Similarly, a lab result of “positive” is meaningless without knowing the threshold used by that specific lab.
- Varying terminologies: Healthcare uses dozens of coding systems (ICD, SNOMED CT, LOINC), each with its own vocabulary. Mapping between them is a minefield. For example, the transition from ICD-9 to ICD-10 was not a simple one-to-one conversion; some codes were split into multiple more specific codes, while others were merged. Mapping these to the highly granular SNOMED CT ontology adds another layer of complexity. Many institutions also use local or proprietary codes that have no direct equivalent in standard terminologies.
- Context dependency: A “diagnosis” in a busy ER is different from one in a controlled clinical trial. The former might be a preliminary assessment, while the latter is a confirmed condition after extensive testing. Losing that context strips away critical information and can lead to flawed conclusions.
- Data quality issues: Missing values, inconsistent granularity (e.g., medication dosage recorded as “one pill” vs. “500mg”), typos in patient identifiers, and inconsistent date formats (‘MM/DD/YY’ vs. ‘YYYY-MM-DD’) make harmonization exponentially harder.
- Administrative barriers: Regulations like HIPAA and GDPR, along with internal institutional policies and data use agreements, create bureaucratic walls around data, making it difficult to even access the data that needs to be harmonized.
The result? Organizations spend up to six months on harmonization, only to find that 30% of their data is unusable. This is a massive waste of resources and a missed opportunity to advance healthcare.
The Challenge of Diverse Data Standards
Multiple data standards exist, but they don’t work together seamlessly.
- HL7 FHIR: A modern standard for making EHR data accessible via APIs, popular in web development.
- OMOP CDM: Designed for observational research, it transforms data into a standardized structure for large-scale population analysis.
- CDISC: The gold standard for clinical trials, providing guidelines for data collection and regulatory submissions.
- openEHR: Focuses on vendor-neutral clinical models using archetypes, popular in Europe and Australia.
- Phenopackets: A schema for exchanging detailed phenotypic and genetic data, crucial for rare disease research.
Each standard has its strengths, but their structural and conceptual differences create significant barriers. A blood test formatted as a FHIR “Observation” resource doesn’t automatically translate to OMOP’s “Measurement” table structure. The mapping requires a deep understanding of both models. For example, you must correctly map the FHIR resource’s subject to the OMOP person ID, the effectiveDateTime to the measurementdate, and the valueQuantity to the valueas_number. Crucially, you must also map the vocabulary, such as converting the LOINC code used in FHIR to a standard concept ID in the OMOP vocabulary. This often requires complex and brittle ETL (Extract, Transform, Load) pipelines that must be maintained and updated constantly. At Lifebit, our platform is built to bridge these gaps without forcing organizations to abandon their existing systems.
The Breakthrough Framework That Open ups Health Data
The real breakthrough in health data harmonization is treating it as a translation problem, not a formatting problem. The most powerful mappings preserve what the data actually means.

A concept-based approach focuses on the underlying idea a piece of data represents. Instead of creating new, context-dependent mappings for every project, you build reusable mappings anchored to a core concept. This allows you to query data from wildly different sources and makes large-scale analysis possible.
Concept vs. Representation: The Key to Accurate Health Data Harmonization
The ISO/IEC 11179 standard provides a powerful framework by distinguishing between what data means and how it’s written down.
- The data element concept is the real-world idea (e.g., “a person’s date of birth”).
- The representation is how it’s expressed (e.g., ‘YYYY-MM-DD’, ‘MM/DD/YYYY’, or as a Unix timestamp).
Harmonizing at the concept level preserves meaning, creating robust mappings that are valid regardless of how different systems encode the data.
The Agency for Healthcare Research and Quality (AHRQ) pioneered this with their Outcomes Measures Framework (OMF). By convening clinical experts to harmonize definitions for complex outcomes like “hospital readmission rates,” “adverse drug events,” and specific conditions like asthma and depression, they created standardized libraries of outcome measures that work in the real world. For each outcome, they established a consensus on the precise clinical definition, identified the specific data elements required to calculate it (e.g., admission dates, discharge status, diagnosis codes), and specified the exact codes from standard terminologies (like ICD and CPT) to identify those elements. For example, their harmonized asthma outcomes work shows how this approach leads to more accurate, reusable mappings that allow different studies to measure the same outcome consistently.
Leveraging Semantics for Scalable Health Data Harmonization
Ontologies and semantic technologies make concept-based harmonization work at scale. Initiatives like the Unified Medical Language System (UMLS) and Mondo Disease Ontology show how shared concepts can align terms across different systems. UMLS, for instance, is a massive metathesaurus that connects hundreds of biomedical vocabularies, creating a unified conceptual space. It’s the engine that allows a system to understand that “Myocardial Infarction” from one vocabulary and “Heart Attack” from another refer to the same clinical concept. At Lifebit, we use FAIR-compliant reference ontologies enriched with vocabularies like SNOMED CT, RxNorm, and LOINC. This knowledge-based strategy helps us identify when “blood tests” and “hematological tests” are conceptually the same, despite different terminology.
The SSSOM framework (Simple Standard for Sharing Ontological Mappings) is a game-changer. It provides a standard way to represent and exchange semantic mappings in a simple, shareable format. Crucially, it goes beyond simple “A equals B” statements by allowing you to enrich mappings with metadata, such as who created the mapping, why it was created (mapping_justification), a confidence score, and the specific rules followed. This metadata makes the mappings transparent, auditable, and reusable, supporting the data provenance that is non-negotiable for regulatory compliance and scientific reproducibility.
Our approach never touches the original data. We perform mappings dynamically, on-the-fly. When source datasets change, the mappings adapt automatically, providing essential flexibility.
| Standard | Primary Use Case | Key Features | Preferred Terminologies |
|---|---|---|---|
| HL7 FHIR | Data exchange, EHR integration, APIs | Resources, RESTful APIs, extensible | LOINC, SNOMED CT |
| OMOP CDM | Observational research, real-world evidence | Standardized tables, concepts, vocabulary, analytics-ready | SNOMED CT, RxNorm, LOINC |
| CDISC | Clinical trials, regulatory submissions | Standardized data collection, management, analysis, reporting | NCIT |
| openEHR | EHR architecture, clinical models, long-term data | Archetypes, templates, vendor-neutral, clinical domain focus | (flexible, often SNOMED CT) |
| Phenopackets | Phenotypic data exchange, rare disease research | Structured representation of phenotypic abnormalities, diagnoses, genes, onset | HPO, Mondo |
Key Steps to Implement a Harmonization Strategy
Implementing a concept-based strategy is a structured, iterative process.
- Define Scope and Goals: What specific research questions are you trying to answer? This step must involve all stakeholders—researchers, clinicians, IT staff, and data scientists—to ensure the goals are clinically relevant, technically feasible, and aligned with the overall objective. Identify the precise data elements needed (e.g., systolic blood pressure, specific medications, diagnosis codes for comorbidities).
- Catalog Your Data Landscape: Document all data sources and the standards, models, and terminologies each one uses (e.g., FHIR, OMOP, CDISC). This includes performing data profiling to understand data quality, completeness, value distributions, and missingness before mapping begins. This initial assessment prevents wasted effort on data that is too sparse or unreliable.
- Establish the Conceptual Model: Determine the underlying concept for each required data element. The role of domain experts (e.g., oncologists for a cancer study, cardiologists for a heart disease study) is non-negotiable here. They are the only ones who can resolve clinical ambiguities and ensure the conceptual model accurately reflects clinical reality. Group similar concepts into clusters.
- Build the Technical Framework: Use standards like ISO/IEC 11179 and SSSOM to represent mappings. This framework should include a metadata repository (MDR) to centrally store, manage, and version the concepts, their representations, and the mappings between them. This creates a single source of truth for all harmonization activities.
- Create and Validate Mappings: Build the connections between source data elements and the central concepts. This is not a one-time task. Validation must be an iterative process with tight feedback loops where clinical experts can review, correct, and refine mappings. Automated quality checks can flag potential inconsistencies or low-confidence mappings for expert review.
- Implement Dynamic Mapping: Integrate the SSSOM mappings into a query federation engine or virtual data model. This allows for on-the-fly harmonization when a query is executed, rather than creating a new, physically transformed and potentially duplicative dataset. This approach preserves the original source data, ensures full data provenance, and reduces storage and maintenance overhead, making the entire process more sustainable.
The Payoff: Real-World Impact of Harmonized Data
When you invest in health data harmonization, you’re not just solving a technical problem—you’re open uping possibilities that can save lives, accelerate breakthroughs, and fundamentally change how we understand human health.

By making diverse datasets interoperable, we gain comprehensive insights from populations we could never study before. Researchers can pool data from multiple institutions, dramatically increasing statistical power. AI models can train on diverse, representative datasets. Public health officials can track disease patterns in real time.
The impact is felt everywhere: clinical research accelerates, AI models become more accurate, patient care improves, and pharmacovigilance becomes more reliable.
At Lifebit, we’ve seen these benefits through our federated platform. When organizations can securely access and analyze global biomedical data without moving it, the pace of innovation increases exponentially. Our Trusted Research Environment (TRE) and Real-time Evidence & Analytics Layer (R.E.A.L.) enable exactly this kind of secure, harmonized collaboration at scale.
Powering Next-Generation Clinical Research
Health data harmonization transforms clinical research. Multi-site studies, which once took over six months just for data preparation, can now be launched almost instantly.
Harmonized data allows researchers to query millions of patient records in minutes. Networks like ACT, PCORnet, and the global OHDSI (Observational Health Data Sciences and Informatics) community leverage common data models to speed up trial recruitment and enable comparative effectiveness research across dozens of health systems.
The benefits are enormous:
- Increased statistical power from larger, combined datasets.
- More reproducible and generalizable findings based on diverse populations.
- Faster study setup, eliminating months of data wrangling.
Consider a hypothetical multi-national study on a new Alzheimer’s drug. Data comes from hospitals in the US (using FHIR), a research database in Europe (using openEHR), and clinical trial data (using CDISC). Without harmonization, this study is impossible. With it, researchers can create a unified cohort by mapping concepts like “cognitive assessment score” (from different neuropsychological tests), “medication history” (from different drug coding systems), and “amyloid-beta biomarker results” (from different lab assays) from all three sources. This allows them to analyze the drug’s effectiveness across diverse genetic backgrounds and healthcare systems, generating evidence that is far more robust and applicable to the global population.
AHRQ’s work on harmonizing outcome definitions, such as their influential harmonized asthma outcomes, enables consistent measurement across studies. For rare diseases, where patients are few and scattered globally, combining data from multiple centers is essential for achieving the statistical power needed to draw life-saving conclusions.
Training Unbiased AI Models at Scale
To build AI that works in the real world, you need harmonized data. It’s that simple.
AI models learn from the data they’re fed. If that data is inconsistent or biased, the AI will be too. A famous example is an AI algorithm for detecting skin cancer that was trained primarily on images of light-skinned individuals. The resulting model performed poorly at identifying melanomas in patients with darker skin, creating a dangerous health inequity. This is a common failure point for healthcare AI.
Health data harmonization solves this by creating consistent, high-quality training datasets from globally representative populations. By harmonizing data from diverse demographic, geographic, and clinical settings, we can ensure the training set reflects the real world, which strengthens the model, making it more robust, generalizable, and equitable.
Privacy is also paramount. With federated learning—an approach Lifebit’s platform is designed for—you can train AI on decentralized data without moving sensitive patient information. This “data-stay” model is critical for overcoming governance and privacy barriers. The harmonization logic (the semantic mappings) can be distributed along with the model to each data source. This allows for local, on-the-fly transformation of the data into a common structure before the model trains on it, all without centralizing the raw data. The model trains locally at each site, and only the aggregated, anonymous model updates are shared. This ensures compliance with HIPAA, GDPR, and other regulations.
The result? Next-generation predictive models that are more accurate, less biased, and applicable across diverse real-world settings. For our biopharma and public health partners, this means faster AI-driven safety surveillance, more accurate clinical trial matching, and better treatment predictions for individual patients.
What’s Next for Health Data: Overcoming Problems to Open up Opportunities
The path to seamless health data harmonization is exciting, but it’s not without its problems. Medical language is complex and context-dependent, making fully automated mapping difficult. Data standards are constantly evolving, requiring continuous maintenance of mappings. Forcing diverse data into rigid structures can also lead to a loss of clinical nuance.
But here’s where it gets exciting. These very challenges are pointing us toward transformative opportunities.
- Smarter AI and Machine Learning: Automated harmonization techniques are reducing manual effort and amplifying the work of human experts. For example, Large Language Models (LLMs) can be trained to suggest potential semantic mappings between different terminologies, which human experts can then quickly validate or correct. This semi-automated approach can drastically speed up the tedious mapping process. Furthermore, Natural Language Processing (NLP) can extract structured data (like diagnoses, medications, and lab values) from unstructured clinical notes, unlocking a vast and rich source of information that is often ignored.
- Global and Cross-Lingual Solutions: As research becomes more international, future frameworks must bridge not just data standards but also languages and cultural contexts. A global pandemic surveillance system, for instance, needs to harmonize data from China (using local standards), Europe (using a mix of standards), and the US (using FHIR). This requires not only mapping data models but also translating and mapping terminologies across languages—for example, mapping a diagnosis described in Mandarin to a universal SNOMED CT concept.
- Dynamic Conceptual Models: Adaptive frameworks are replacing static, brittle mappings. Tools like LinkML make it easier to define and maintain flexible data schemas that can evolve with medical knowledge. LinkML is a modeling language that allows you to define a schema (a blueprint for your data) and then automatically generate different consistent representations from it, such as JSON-Schema, Python data classes, and even SSSOM mapping files. This makes the conceptual model the single source of truth and ensures consistency as it evolves.
- Community-Driven Validation: Collaborative, open review of mappings is becoming the norm. Platforms like GitHub allow mapping sets to be shared, versioned, and collaboratively improved by the global research community, much like open-source software. This fosters transparency, builds trust, and creates a shared, high-quality resource that benefits everyone.
- FAIR Data Principles as the North Star: Making data Findable, Accessible, Interoperable, and Reusable is fundamental. This requires transparent documentation, rich metadata about data provenance, and clear licensing to build confidence in the entire system and encourage data sharing.
- Accelerating Global Collaboration: Governments, healthcare organizations, and researchers are increasingly working together on universal standards. Initiatives like the Global Alliance for Genomics and Health (GA4GH) and AHRQ’s Outcomes Measures Framework provide a blueprint for large-scale, consensus-driven harmonization.
The vision is coming into focus: a centralized mapping system based on standardized concepts, where the rich diversity of health data from different standards can be woven together into a coherent, queryable resource. This doesn’t mean forcing everyone to use the same standard—it means creating the bridges that let different standards communicate effectively.
At Lifebit, we’re building these bridges every day. Our federated platform doesn’t just connect data—it understands it, preserving meaning while enabling analysis at scale. The future of data interoperability isn’t about eliminating diversity; it’s about celebrating it while ensuring that valuable insights aren’t lost in translation.
Frequently Asked Questions about Health Data Harmonization
What is the main goal of health data harmonization?
The main goal of health data harmonization is to make data from different sources (e.g., hospitals, clinical trials, genomics databases) comparable and interoperable. It resolves differences in format, structure, and meaning so the data can be analyzed together, enabling large-scale research and better patient care without losing the original clinical context.
How does harmonization differ from standardization?
Standardization imposes a single, uniform format on all data, like converting everything to the OMOP CDM. It’s a “one size fits all” approach.
Harmonization is more flexible. It creates a common framework that allows different standards (like FHIR, CDISC, and OMOP) to coexist while being understandable in relation to each other. It builds bridges between data models rather than forcing conformity, allowing for easier integration without requiring partners to overhaul their systems.
What is a “concept-based” approach?
A concept-based approach focuses on harmonizing the underlying meaning (the ‘concept’) of data, not just its label or format (the ‘representation’). For example, it understands that different ways of recording “blood pressure” refer to the same clinical idea. This creates more accurate and reusable mappings that don’t break when data formats change, ensuring the “what” of the data remains consistent across systems.
Conclusion: Unifying Data to Revolutionize Healthcare
Fragmented health data is holding back progress. Researchers waste months on data wrangling, AI models learn from biased information, and patients wait for personalized care that remains out of reach. The cost of inactiondelayed breakthroughs and missed opportunitiesis too high.
At Lifebit, we’ve seen that concept-based, semantic harmonization is the solution. By focusing on the underlying meaning of data, we can break down silos. Our work with frameworks like SSSOM proves you can create accurate, reusable mappings for dynamic harmonization that respects data integrity while open uping powerful analysis.
This unified approach transforms scattered information into coherent insights. It accelerates research, trains fairer AI models, and enables the kind of real-time evidence generation that can catch safety signals before they become tragedies.
Most importantly, it improves patient outcomes on a global scale. When data flows freely between systems while remaining secure and private, every patient benefits from the collective knowledge of healthcare worldwide.
Our federated platform provides the advanced tools you need to implement these strategies securely across global and multi-omic datasets. Whether you’re in biopharma, government, or public health, we’re here to help you open up the full potential of your health information. Our Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence & Analytics Layer) work together to deliver the real-time insights, AI-driven safety surveillance, and secure collaboration that modern healthcare demands.
The future of medicine depends on our ability to see the whole picture, not just isolated pieces. Find out how to unify your health data with our federated solutions and join us in building that future.

