Genome Browser: Analysing Genomic Data Without Bioinformatics Skills

As sequencing technologies become more performant year over year, three challenges for the research community arise: 1) astronomical quantities of data are generated, 2) data files are more complex, and 3) more computational expertise is required to analyse and manipulate sequencing files for genomics data analysis without coding. This need for improved methods of genomics data analysis without coding underscores the importance of accessible tools.
Moreover, as it becomes more affordable to implement sequencing in biomedical research, scientists with varying levels of programming skills are required to interact with the same complex data files, including bench scientists and clinicians. Although these individuals have knowledge of the science behind the data, they frequently lack the computing skills to manipulate the data files and perform genomics data analysis without coding to derive actionable insights.
Researchers can leverage the power of genomics data analysis without coding to quickly interpret complex datasets.
These advancements in visualization tools are crucial for effective genomics data analysis without coding.
Utilizing methods for genomics data analysis without coding has become essential in modern research.
Good news! The advent and refinement of visualisation tools has allowed scientists of all programming levels to unearth the valuable insights that lie in these complex file types, which can consist of thousands to billions of data points. One such tool is the Integrative Genomics Viewer (IGV), a high-performance visualisation tool for the interactive exploration of large genomic files.
Using a genome browser simplifies genomics data analysis without coding, making it accessible to all researchers.
The Integrative Genomics Viewer (IGV) genome browser: a picture is worth a thousand words & lines of code 😉
Unlocking Insights: Genomics Data Analysis Without Coding
Genome browsers, such as IGV, turn very large sequencing files into easy-to-understand visualisations that contain both coverage and genotypic information. Essentially, they are an alternative to processing the data algorithmically through Python, R or other scripts. By using a genome browser you save yourself the trouble of writing hundreds to thousands of lines of code!
Furthermore, understanding genomics data analysis without coding can enhance collaborative efforts in research.
IGV’s graphical interface enables the real-time intuitive exploration of large-scale genomic datasets, and supports heterogeneous data types including array-based and next-generation sequencing data and genomic annotations (i.e. BAM, VCF(.gz), BED, BW, BEDGRAPH(.gz)).
Exploring aligned genomic data through visualisations
Start your journey in genomics data analysis without coding today with IGV!
With IGV on CloudOS, genomics data analysis without coding has never been easier.
The interactive exploration of the genome aligned against a reference can be used for a variety of different research applications ranging from simple SNP querying to more elaborate epigenetic studies exploring the highly regulated regions. As such, IGV is a great tool for bench scientists and clinicians alike, who have a strong understanding of the biology behind their hypotheses, but lack the computational skills to carry out the deep exploration of their samples.
The main uses of IGV genome browser include, but are not limited to:
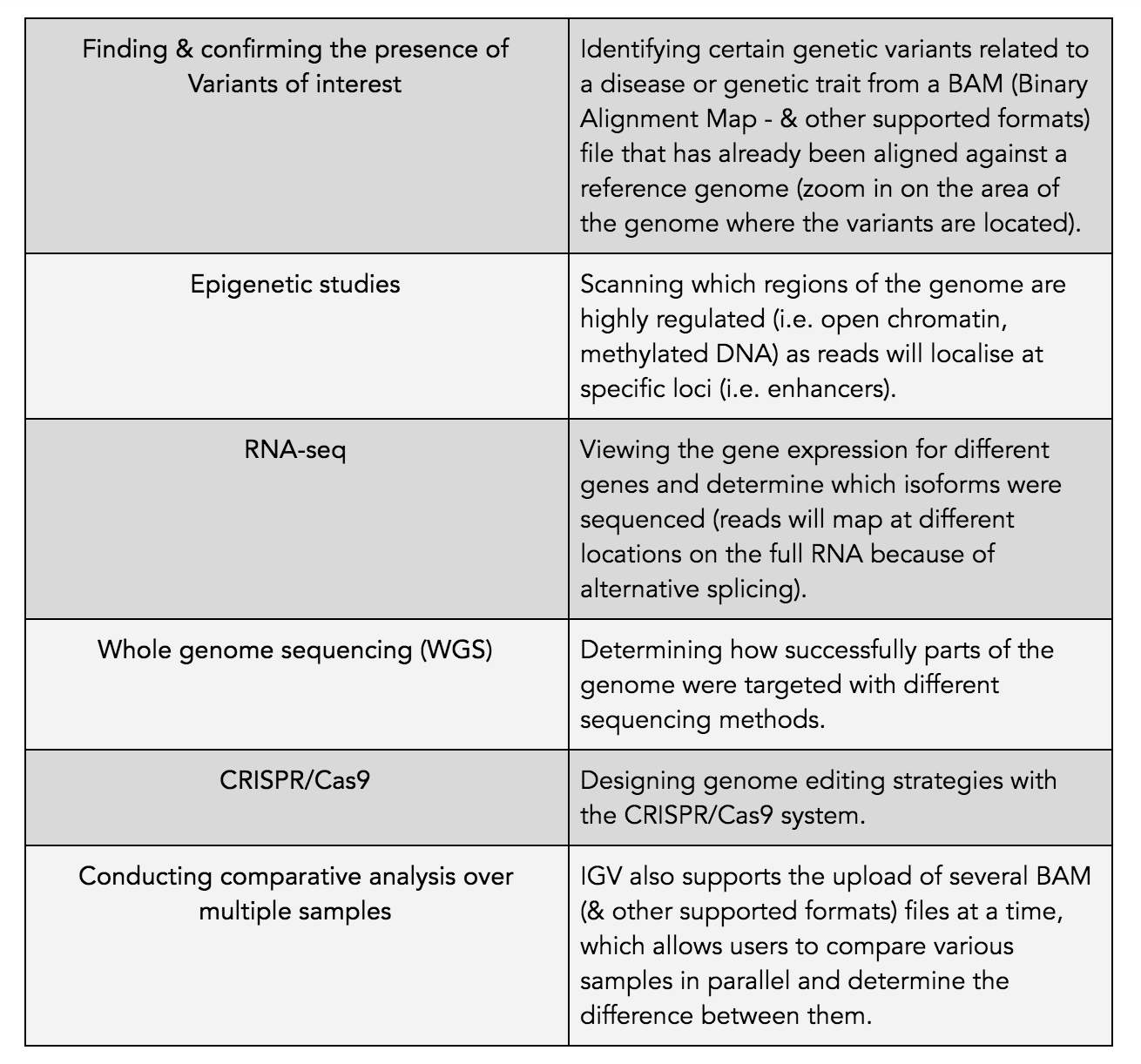
The IGV genome browser is not only limited to scientists lacking programming skills, as seasoned bioinformaticians often use IGV for quality control of their samples. This enables the inspection of data right after secondary analysis, while informing tertiary analysis. Furthermore, IGV can also generate intuitive graphics for publication purposes.
Start now with IGV over CloudOS for free
And for the greater news! The IGV genome browser is now full integrated on Lifebit’s CloudOS platform. This means that you can start using IGV right away and for free, forever, alongside many other researchers and clinicians!
Being able to access IGV directly through CloudOS allows you to:
- Apart from uploading files, directly select files from your Cloud AWS S3 bucket: this saves time and mitigates the risk of transferring sensitive data files out of your environment,
- Efficiently collaborate and visualise the same files, while forgoing time-consuming data transfers, and to
- Create informative visualisations to confirm research findings.
How does it work, you ask? It only takes 3 clicks of the mouse, it’s that simple!
- Simply upload or select from your AWS S3 bucket the BAM files (or any other supported file type) you wish to visualise.
- Zoom in on your area(s) of interest in the genome and visualise away!
- Optional: save your visual analysis as an SVG file for future reference.
We encourage you to explore the possibilities of genomics data analysis without coding and share your experiences.
IGV can also generate intuitive graphics for publication purposes. Start now with IGV over CloudOS for free. Accessing IGV directly through CloudOS allows you to analyze genomics data without requiring bioinformatics skills through Lifebit’s CloudOS platform.
Now that you have a new superpower on Lifebit CloudOS, we would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!

