From Data to Discovery: Navigating the Biomarker Landscape

From Raw Data to Life-Saving Drugs: The Digital Biomarker Revolution
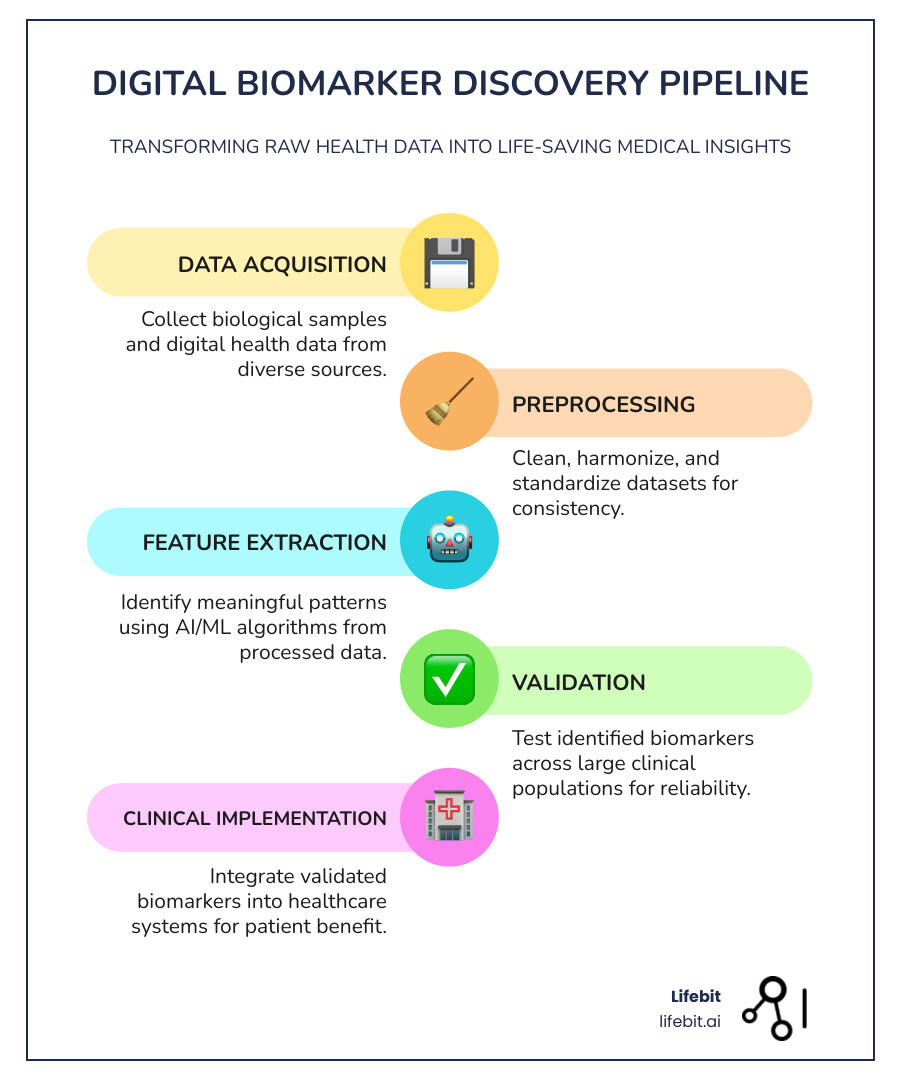
A biomarker findy pipeline systematically transforms raw health data into validated medical insights. It’s a multi-stage process designed to identify, validate, and clinically apply measurable biological indicators that can predict, diagnose, or monitor disease.
Core Pipeline Stages:
- Data Acquisition – Collecting biological samples and digital health data.
- Preprocessing – Cleaning, harmonizing, and standardizing datasets.
- Feature Extraction – Identifying meaningful patterns with AI/ML.
- Validation – Testing biomarkers across large clinical populations.
- Clinical Implementation – Integrating validated biomarkers into healthcare.
The stakes are immense. Cardiovascular diseases are the world’s leading killer, yet most biomarker candidates fail before reaching clinical use. The problem isn’t a lack of candidates from ‘omics’ tech; it’s the validation bottleneck where promising findies face the harsh reality of clinical application.
Digital biomarkers are changing this. By changing data from smartphones and wearables into continuous health indicators, we can monitor patients 24/7, detect disease earlier, and personalize treatments like never before. But the challenge is staggering: only 0-2 new protein biomarkers achieve FDA approval per year across all diseases. This gap costs billions in failed R&D and delays life-saving treatments.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit. For over 15 years, I’ve built computational tools for precision medicine and biomarker findy pipelines in federated data environments, focusing on breaking down the data silos that stall validation and moving findies into the clinic.
What Is a Digital Biomarker? (And Why It’s a Game-Changer)
A digital biomarker is an objective health indicator derived from data collected by digital devices like smartwatches, smartphones, or other biometric monitoring technologies (BioMeTs). Instead of a one-time blood test, imagine continuous data streams on heart rate, sleep patterns, and activity levels.
This ushers in an era of proactive healthcare. Continuous monitoring detects subtle changes that signal disease onset long before symptoms appear, enabling earlier intervention and personalized management. For example, abnormal heart rate patterns from a wearable can flag an impending cardiovascular event. This real-time data flow is a massive leap from the episodic snapshots of traditional clinical visits. Digital biomarkers also enrich Patient-Reported Outcomes (PROMIS) with objective data, creating a more accurate picture of a patient’s health and treatment response.
The $2M Problem: Why Traditional Biomarkers Keep Failing
Traditional biomarkers, while foundational, have limits. They are typically single-point, invasive measurements (blood draws, biopsies) that are expensive and inconvenient. These snapshots often miss the dynamic changes of early-stage disease.
Consider the scale: Cardiovascular Diseases (CVDs) are the top cause of death globally, yet traditional ECGs can miss preclinical signs of heart failure. The process for finding traditional biomarkers is also broken. Thousands of candidates are identified, but few are validated. The “tar pit” is the verification stage, where developing an ELISA for just one candidate can cost up to $2 million, take over a year, and still have a high failure rate. This bottleneck, combined with regulatory problems and data-sharing mistrust, means many promising biomarkers never help patients
Build a Biomarker Pipeline That Works: The 3-Stage Blueprint
A high-impact biomarker findy pipeline turns raw health data into clinical breakthroughs. Unlike pipelines that get stuck in research limbo, a successful one is end-to-end, modular, and built on FAIR principles (Findable, Accessible, Interoperable, Reusable). This ensures that data, tools, and algorithms are findable and reusable, separating scalable solutions from interesting but unproven research.

Stage 1: From Messy Raw Data to Clean, Standardized Insights
Breakthroughs start with data from wearable devices, smartphones, and EHRs. But this raw data is messy and inconsistent—different devices and systems use unique formats and algorithms. The first critical step is data cleaning and data harmonization.
Without proper standardization, comparing heart rate data from different smartwatches is impossible. This is why standardized data formats like The Brain Imaging Data Structure (BIDS) standard are crucial, especially for complex EEG data. Standards ensure that data is comparable and reproducible across studies. Preprocessing involves technical steps like signal noise reduction (e.g., removing artifacts from EEG data) to create a rock-solid foundation for analysis.
Stage 2: Using AI to Find Hidden Patterns (Without the Black Box)
This stage turns clean data into insights through feature engineering and signal processing. Here, we extract hidden patterns—like Alpha Peak Frequency (APF) from EEG or heart rate variability from wearables—that could become life-saving biomarkers.
AI/ML models excel at spotting subtle patterns humans would miss. However, many models are “black boxes,” making predictions without explaining their reasoning. This is why Explainable AI (XAI) is essential, not optional. For a doctor or regulator to trust an AI-driven biomarker, they must understand why it made a specific prediction. Interpretability builds trust and is critical for clinical acceptance. Our process integrates XAI from the start, ensuring our digital biomarkers are predictive, understandable, and clinically actionable.
Stage 3: The Ultimate Test—Clinical Validation at Scale
This is where most biomarker projects fail. Clinical validation requires rigorous proof that a biomarker works in the real world, not just in a controlled lab. It must demonstrate reliability, sensitivity, and specificity across large, diverse populations through extensive statistical analysis against established clinical endpoints.
Reproducibility is non-negotiable. If different labs cannot get the same results, the biomarker is clinically useless. Validation requires large-scale datasets, like the LEMON dataset (213 healthy participants) and TDBRAIN dataset (1,274 participants), to confirm a biomarker’s utility across diverse groups. As detailed in Building better biomarkers in neuroimaging, this rigorous process is what separates interesting findings from tools that actually save lives.
Why 99% of Biomarker Pipelines Fail (And How to Fix Them)
Developing a successful biomarker findy pipeline is notoriously difficult. The path is littered with challenges: data silos trap valuable information in isolated systems, standardization gaps make combining datasets impossible, and a reproducibility crisis undermines scientific trust. On top of this, navigating ethical considerations and data privacy is paramount. These problems often feel overwhelming, but they are solvable with the right approach.
The 5 Pitfalls That Kill Biomarker Projects
Most promising biomarker projects stall for predictable reasons. Avoiding these common culprits is key to success.
- Lack of Large Datasets: Findings from small cohorts often fail to generalize across larger, diverse populations. Without massive, representative data, a biomarker’s utility remains unproven.
- Analytical Variability: Different teams using slightly different methods (e.g., processing EEG data at different frequencies) can produce conflicting results, creating chaos and invalidating comparisons.
- Ethical and Privacy Barriers: Fear of data misuse and a lack of bulletproof privacy safeguards can halt collaboration, severely limiting the scope and scale of research.
- Missing Governance Frameworks: Without clear rules on data ownership, access, and use, even willing partners struggle to collaborate effectively.
- The “Small n, Large p” Problem: In ‘omics’ research, we often have thousands of potential features (genes, proteins) but a small number of patients, making it statistically difficult to find meaningful signals.
The Open-Source Fix: How DBDP Accelerates Findy
Open-source initiatives like the Digital Biomarker Findy Pipeline (DBDP) offer a powerful solution. The DBDP on GitHub is an open-source project promoting toolkits, reference methods, and community standards to overcome common development problems.
The DBDP is built on FAIR principles (Findable, Accessible, Interoperable, Reusable), creating an ecosystem where data, algorithms, and code are transparent and verifiable. This directly tackles the biggest challenges:
- Standardization: Modular frameworks reduce analytical variability.
- Reproducibility: Open-source code enables transparent, verifiable methods.
- Collaboration: Shared resources like DISCOVER-EEG, an automated pipeline for EEG data, allow teams to build on proven foundations instead of reinventing the wheel.
- Accessibility: Resources like the Digital Health Data Repository (DHDR) provide sample datasets, while dbdpED offers education on using mHealth data.
Operating under an Apache 2.0 License and partnering with organizations like Open mHealth, the DBDP fosters a collaborative spirit essential for moving biomarkers from findy to clinical practice.
Digital Biomarkers in Action: Real-World Clinical Wins
The promise of digital biomarkers is already a reality, shifting medicine from reactive treatment to predictive, personalized care.
Predicting Heart Attacks: How Digital Biomarkers Are Saving Lives
Cardiovascular diseases are the world’s #1 killer, but biomarker findy pipelines are changing that. Continuous data from wearables is revolutionizing early detection. Advanced digital biomarkers can identify subtle heart rate variability patterns that signal trouble weeks before symptoms appear. Novel ECG biomarkers are indicating CVD risk long before traditional methods.
Remote patient monitoring allows cardiologists to track heart health 24/7, providing early alerts for chronic conditions. This leads to better outcomes, fewer ER visits, and reduced hospital readmissions. Integrated predictive analytics allow doctors to forecast disease progression and adjust treatments proactively, creating a real-time, evolving care plan.
Decoding the Brain: New Biomarkers for Depression and Alzheimer’s
Digital biomarkers are providing the first objective windows into complex brain conditions like psychiatric disorders and Alzheimer’s disease. Electroencephalography (EEG)—non-invasive, portable, and cost-effective—is a key tool. Automated pipelines like DISCOVER-EEG process brain activity to identify biomarkers for depression subtypes, moving beyond subjective assessments.
Large-scale validation is proving their power. Studies using datasets like LEMON (213 healthy participants) and TDBRAIN (1,274 participants with psychiatric disorders) are revealing significant patterns. For example, analysis of the LEMON dataset showed strong evidence that older individuals have a lower Alpha Peak Frequency (APF), a potential biomarker for brain aging. These reproducible brain-wide association studies are accelerating our understanding of neurological and mental health, creating objective indicators to guide diagnosis and track recovery.
Your Top 3 Questions About Biomarker Findy, Answered
What is the difference between a biomarker and a digital biomarker?
A traditional biomarker is a static snapshot of health, like a protein level in a blood test or a spot on an MRI. It’s typically invasive, expensive, and captures only a single moment in time.
A digital biomarker is a continuous video stream of your health, derived from data collected by digital devices like smartphones and wearables. It provides real-world insights over time, tracking patterns in gait, heart rhythm, or sleep. This allows for the detection of subtle changes that signal disease long before a traditional test would.
What are the FAIR principles?
FAIR stands for Findable, Accessible, Interoperable, and Reusable. It’s a set of guiding principles for data and code that is crucial for successful biomarker findy pipelines.
- Findable: Data and tools have unique identifiers and rich metadata so they can be easily finded.
- Accessible: Data can be retrieved by authorized users through open, standardized protocols.
- Interoperable: Data uses shared formats and vocabularies, allowing different datasets to be combined and analyzed together.
- Reusable: Data is well-documented with clear licensing so it can be used in future studies.
FAIR principles prevent researchers from reinventing the wheel and accelerate medical breakthroughs.
What is the role of Explainable AI (XAI) in biomarker findy?
In healthcare, accuracy isn’t enough. Clinicians and regulators need to understand why an AI model makes a prediction. Explainable AI (XAI) makes AI decisions transparent and interpretable, solving the “black box” problem.
Instead of just a risk score, XAI shows which features (e.g., sleep patterns, activity levels) drove the prediction. This builds clinical trust, is often required for regulatory approval (e.g., by the FDA), and helps validate the biological plausibility of a new biomarker. Without XAI, even the most accurate AI-derived biomarkers will fail to gain clinical adoption.
The Future of Biomarker Findy is Federated. Don’t Get Left Behind.
The journey from sensor data to medical insight is shifting healthcare from “wait and treat” to “predict and prevent.” But the most powerful algorithms are useless if they’re locked in data silos. The future of the biomarker findy pipeline is not just digital—it’s federated and collaborative.
Federated learning solves the data-sharing problem. Instead of moving sensitive patient data, we bring the algorithms to the data. Insights are shared, but the raw data stays secure and private. This approach, combined with real-time evidence generation, accelerates the entire pipeline from decades to months. We can validate biomarkers across diverse, global populations simultaneously, spot safety signals faster, and adapt as new data streams in.
At Lifebit, our platform is built to break down these barriers. Our Trusted Research Environment (TRE) enables secure analysis of sensitive data, while our Trusted Data Lakehouse (TDL) harmonizes disparate sources like EHRs, wearables, and genomics. The R.E.A.L. (Real-time Evidence & Analytics Layer) delivers the insights that matter, when they matter.
By combining open-source initiatives like the DBDP with secure federated platforms, we create a collaborative ecosystem where researchers build on each other’s work and pharma can validate biomarkers globally without moving a single patient record. The bottlenecks of the past—small datasets, irreproducible results, siloed information—are being solved.
The question isn’t if federated biomarker pipelines will transform healthcare, but whether you’ll be part of it.
Learn how federated data analysis is powering the future of research and find out how your organization can lead this revolution in precision medicine.

