The NIAID Data Ecosystem: Connecting Biomedical Research Through Federation

End Data Roadblocks—Accelerate Infectious Disease Research Now
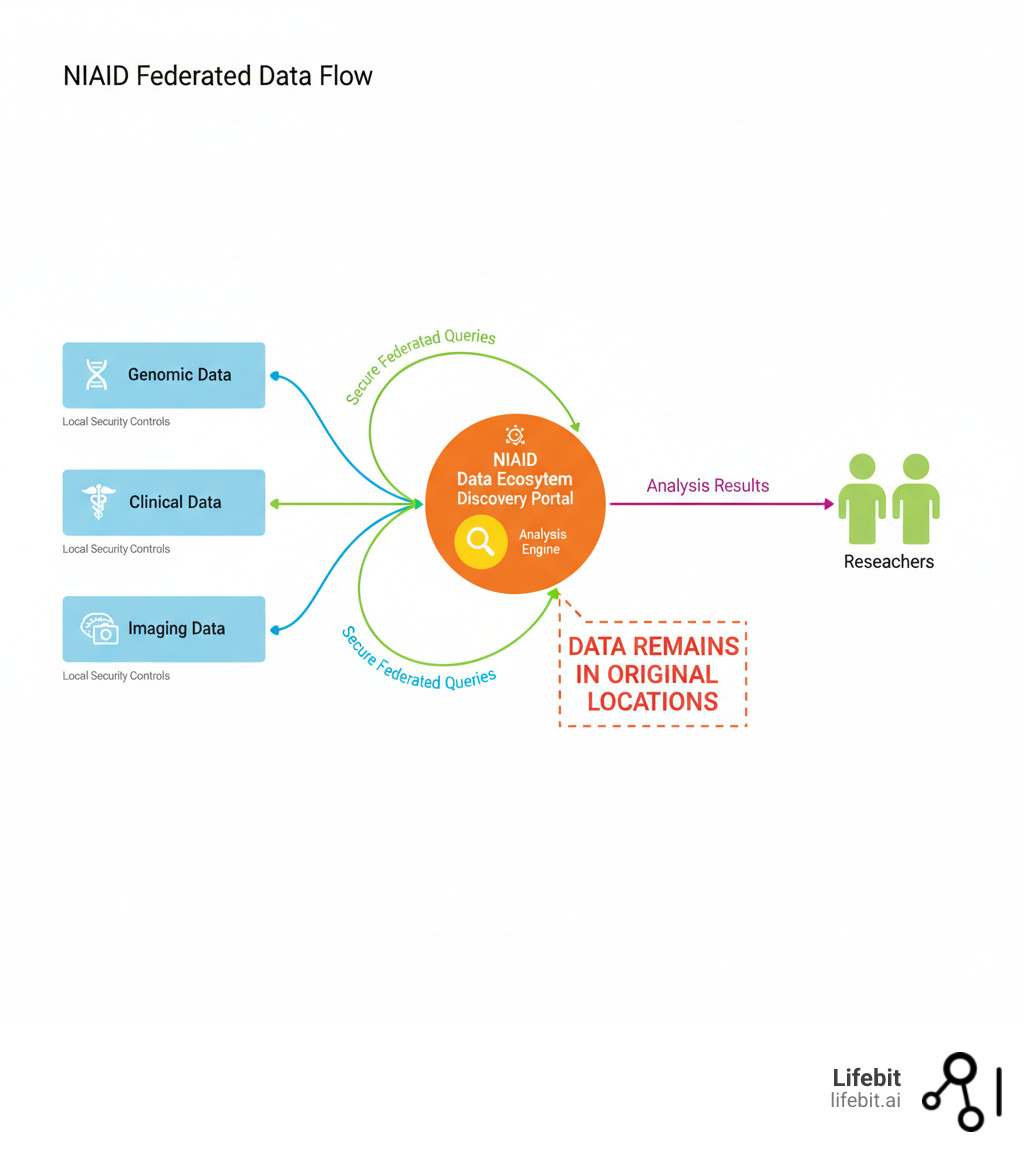
The federated data ecosystem niaid is a secure, distributed network that allows researchers to find, access, and analyze infectious disease data from multiple repositories without moving it. This keeps data owners in control while accelerating research on COVID-19, AIDS, tuberculosis, malaria, and emerging threats.
Key facts about the NIAID Data Ecosystem (NDE):
- Investment: A $7.5M contract was awarded to build the ecosystem
- Purpose: Speed development of diagnostics, therapeutics, and vaccines
- How it works: Data stays local; researchers search across repositories from a single interface
- Security: Data owners maintain control of storage, security, and management
- Access: Through the NIAID Data Ecosystem Findy Portal
- Focus: Infectious and immune-mediated diseases, with emphasis on pandemic preparedness
The COVID-19 pandemic exposed a critical flaw: over 80% of vital research data is locked away. In a health crisis, researchers waste months navigating data silos and security protocols when every day counts.
The NIAID Data Ecosystem fixes this. It’s a federated network where data stays protected by its owners, but researchers can analyze it from one place. No more data transfer delays, security nightmares, or missed opportunities to save lives.
As Maria Chatzou Dunford, CEO of Lifebit, I’ve spent over 15 years building platforms that break down these barriers. From developing Nextflow to creating federated networks for public health and pharma, my work has focused on solving these challenges. This guide explains how NIAID’s federated approach works and why it’s replacing outdated data sharing models in biomedical research.
Why Federation Crushes Centralized Data: Security, Speed, and Control
Imagine you’re a researcher racing to understand a new disease variant. The genomic, clinical, and immune data you need is scattered across dozens of hospitals and research centers globally. Getting access isn’t just a hurdle; it’s the main obstacle to discovery.
Traditional data sharing, often called Collaborative Data Sharing (CDS) or the centralized model, requires physically moving sensitive patient data to a central server. This process is a logistical and legal nightmare, bogged down by months, or even years, of negotiating Data Use Agreements (DUAs), navigating institutional review boards (IRBs), and overcoming immense technical challenges related to data transfer and storage. By the time you finally get access, the science has often moved on, and the public health opportunity may have been missed. This old-school approach is a paradigm of bottlenecks, security vulnerabilities, and prohibitive costs.
The hard truth is that over 80% of biomedical data remains locked away in institutional silos. This isn’t due to a lack of willingness to collaborate, but because institutions are rightly hesitant to relinquish control and move sensitive patient information to third-party servers. The risks of a data breach, the complexities of cross-jurisdictional legal compliance (like GDPR in Europe and HIPAA in the US), and the sheer cost of transferring petabytes of data make centralization an unworkable model for large-scale research.
The federated data ecosystem niaid flips this broken model on its head. Instead of moving data to the analysis, it moves the analysis to the data. Sensitive information stays securely behind institutional firewalls, under the complete control of the local data owner. Only the non-sensitive, aggregated results of the analysis or updated model parameters travel back to the researcher. It’s like sending a secure, vetted recipe to different kitchens instead of shipping all their precious, locally-sourced ingredients to one central location. The ingredients never leave home; you only see the finished dish.
This model is essential for modern biomedical research. Our experience building federated platforms for government agencies and top pharmaceutical companies shows it enables critical collaboration that would otherwise be impossible. Institutions that would never agree to upload their raw data to an external cloud are willing and active participants in federated analyses. Our Federated Data Sharing Complete Guide walks through exactly how this paradigm shift happens and why it’s rapidly becoming the gold standard for sensitive research.
See Federation in Action: Real-World Biomedical Impact
Federated Learning (FL) demonstrates this approach at its most powerful, particularly for training artificial intelligence models. Imagine an AI model that needs to learn to detect a disease from medical images stored at ten different hospitals. In a federated setup:
- Initialization: A central server initializes a global AI model and sends a copy to each of the ten hospitals.
- Local Training: Each hospital trains the model exclusively on its own local data, behind its own firewall. No patient data is ever exposed or moved.
- Secure Update: After a round of training, each hospital sends back only the mathematical updates to the model (the ‘weights’ or ‘gradients’), not the data used to create them. These updates are abstract numerical representations of what the model learned.
- Aggregation: The central server securely aggregates these updates (using methods like Federated Averaging or FedAvg) to create an improved, more intelligent global model.
- Iteration: This improved global model is sent back to the hospitals for another round of local training. This cycle repeats, with the model becoming progressively more accurate by learning from the collective knowledge of all institutions without ever seeing their raw data.
The result is a highly accurate model trained on a vast and diverse dataset, all while ensuring patient privacy and institutional control. A landmark study in Nature Communications validated this, finding that federated learning models across 10 institutions achieved 99% of the accuracy of models trained using a traditional centralized approach—but without sharing a single patient record. That’s a breakthrough for collaborative science.
This isn’t just theoretical. The Federated Tumor Segmentation (FeTS) platform enabled 71 sites across 6 continents to collaborate on glioblastoma research using data from 6,314 patients. This global effort, which would have been impossible with centralized models due to data privacy and ownership concerns, developed a state-of-the-art machine learning model for detecting tumor boundaries. Similar federated initiatives are accelerating research on Alzheimer’s disease, COVID-19 drug efficacy, and the diagnosis of rare genetic disorders, making previously impossible research feasible. We explore these applications in our guide on Federated Learning in Healthcare.
Why Federation Wins: Key Advantages Over Old-School Data Sharing
The benefits of federation are transformative, creating a clear case for its adoption in sensitive research.
Stronger Security: Patient data never leaves its home institution’s secure environment. By only sharing aggregated, non-sensitive results or abstract model parameters, the attack surface is drastically reduced. This eliminates the risk of a catastrophic breach at a central data repository. Furthermore, techniques like differential privacy can be applied during local training to add mathematical noise, making it impossible to reverse-engineer an individual’s data from the model updates.
Real Data Sovereignty: Institutions maintain complete and uninterrupted control over their data assets. They manage their own security, access controls, and compliance. This builds the trust necessary for collaboration, as the promise that “your data never leaves” is a verifiable technical guarantee, not just a contractual one.
Effective Scaling: Analytical power grows exponentially as more institutions join the network, without the spiraling costs of building and maintaining a massive central data warehouse. This is crucial for petabyte-scale genomics and imaging data, as our work on Federated Architecture in Genomics demonstrates.
Explosive Diversity: By removing the friction of data transfer, federation allows smaller clinics, international partners, and institutions serving underrepresented populations to participate in cutting-edge research. This dramatically increases the diversity of the data, leading to more robust, generalizable, and equitable AI models that work for everyone, not just the populations represented in a few large academic centers.
Significant Cost Reduction: Federation eliminates the enormous costs associated with data egress (transfer fees from cloud providers), redundant central storage, and the personnel needed to manage a centralized repository. It leverages existing infrastructure, making large-scale collaboration financially sustainable.
Massive Speed Increase: Researchers can begin analyzing data almost immediately, without waiting for lengthy data transfer and legal processes. This compresses research timelines from years to weeks or even days. In a pandemic, this speed directly translates to saving lives.
| Feature | Centralized Data Sharing (CDS) | Federated Data Ecosystem |
|---|---|---|
| Security | High risk, data moved to central location, single point of failure. | Data stays local, reduced risk, distributed security, improved privacy. |
| Scalability | Challenging with large numbers of collaborators, high infrastructure costs. | Highly scalable, leverages existing infrastructure, easier to add new data sources. |
| Data Sovereignty | Data owners relinquish direct control once data is centralized. | Data owners retain full control and ownership of their data. |
| Compliance | Complex, as centralized data must meet all regulations of all contributing sources. | Simplified, as each institution ensures local compliance. |
| Cost | High for storage, transfer, and management of central repository. | Lower, as it leverages distributed resources and reduces transfer needs. |
| Data Diversity | Limited by willingness to share, potential for bias from centralized collection. | Maximized, as more institutions can participate without moving sensitive data. |
The real difference is what becomes possible. Projects once blocked by institutional, legal, or financial barriers can now proceed smoothly. Collaborations that took years to set up can now happen in months. This is why the federated data ecosystem niaid represents a fundamental shift, making federation the new, undisputed standard for sensitive data research at scale.
NIAID’s $7.5M Bet on Federation: How to Use Its Data Ecosystem
To tackle the data access crisis head-on, the National Institute of Allergy and Infectious Diseases (NIAID) made a bold move, investing $7.5M to build a next-generation platform. The result is the federated data ecosystem niaid—officially known as the NIAID Data Ecosystem (NDE)—a system designed to fundamentally change how infectious disease research is conducted.
This isn’t a typical government IT project. It’s a dynamic partnership between leading technology providers, data stewards, and the research community, all focused on a single, urgent goal: giving scientists secure, seamless, and rapid access to the world’s most critical health data.
The NDE is explicitly not a centralized data commons. Instead of hoarding data in one place, it empowers data owners to retain full control over their data’s storage, security, and governance. The ecosystem provides the connective tissue—the shared infrastructure and standards—to allow researchers to search distributed sources and analyze data where it lives.
The technical backbone of this system is the Data Ecosystem Framework (DEF). The DEF is an open-source, modular gateway that creates secure, interoperable connections between researchers and disparate data repositories. It acts as a universal translator, using standardized APIs and metadata schemas to let a researcher’s query or analysis tool interact with multiple data sources as if they were a single, integrated resource, all while respecting the local security and access policies of each data owner.
What Data Is at Your Fingertips?
The federated data ecosystem niaid is laser-focused on data critical to understanding, treating, and preventing infectious and immune-mediated diseases. This includes data for emerging threats like COVID-19 and mpox, as well as persistent global challenges like AIDS, tuberculosis, and malaria.
The NDE offers a full spectrum of biomedical data types, making multi-modal analysis possible. This includes:
- Genomic and Omics Data: Raw sequence data (WGS, WES), transcriptomics (RNA-Seq), proteomics, and metabolomics from pathogens and human hosts.
- Clinical Data: De-identified electronic health records (EHRs), clinical trial results, patient demographics, comorbidities, and treatment outcomes.
- Immunological Data: Immune phenotyping results from flow cytometry (FACS), cytokine profiles, antibody titers, and T-cell response assays.
- Imaging Data: Medical images such as chest X-rays and CT scans for respiratory diseases or microscopy images for cellular-level analysis.
NIAID has cultivated an impressive collection of Resource Catalogs and Dataset Repositories that are part of this ecosystem. The beauty of federation is that these invaluable resources remain managed and controlled by their expert creators but become findable and usable through a single, unified portal. You can explore the breadth of available data through the NIAID Data Ecosystem Discovery Portal.
Tools That Boost Discovery and Analysis
Finding data is just the beginning. The NDE’s real power lies in the sophisticated tools it provides to analyze that data securely and reproducibly.
The NIAID Data Ecosystem Discovery Portal is your starting point. This single interface leverages indexed metadata from all participating repositories, allowing you to run complex searches across NIAID’s entire diverse data landscape at once. This replaces the tedious and inefficient process of visiting dozens of individual repository websites.
Once you find relevant data and receive access from the data owner, the ecosystem provides secure cloud analysis environments, often referred to as Trusted Research Environments (TREs). A TRE is a highly controlled, locked-down virtual workspace in the cloud. It allows approved researchers to work with sensitive data using powerful computational tools, but prevents the raw data itself from being downloaded or moved. Key features include strict controls on data ingress and egress, comprehensive audit trails of all actions, and a secure perimeter that isolates the analysis from the public internet. At Lifebit, we’ve perfected these Trusted Research Environments, which are the cornerstone of enabling secure, collaborative, and cutting-edge research at scale.
The NDE also champions reproducible, shareable workflows. Science that cannot be reproduced is not reliable. To solve this, the ecosystem supports the use of containerization technologies like Docker and workflow languages like Common Workflow Language (CWL) and Nextflow. These tools allow researchers to package their entire analysis pipeline—including the code, dependencies, and parameters—into a portable, executable object. This ensures that another researcher can re-run the exact same analysis on the same data and get the exact same result, which is essential for validating findings and building upon previous work.
NIAID is actively expanding the ecosystem’s capabilities through new funding opportunities. They seek software that integrates seamlessly with the NDE by being indexable, exposing FAIR metadata via APIs, and adhering to the metadata schema for computational tools. This forward-looking strategy ensures that new AI and ML tools can easily plug into the ecosystem, bringing the latest analytical power securely to the data’s source. Lifebit’s platforms are built from the ground up for this kind of interoperability, making us a key partner in the federated data revolution.
NIH’s Secret Weapon Against the Next Pandemic: The NIAID Ecosystem
COVID-19 exposed a harsh truth: our global scientific infrastructure was not ready. Data silos, proprietary formats, and slow, bureaucratic sharing processes cost us precious time in a crisis where every day mattered. The federated data ecosystem niaid emerged directly from this wake-up call as a vital part of the NIH’s mission to transform our national response to health emergencies.
The NIH leadership realized that pandemic preparedness requires robust, flexible, and scalable data infrastructure to be in place before a crisis hits. The NDE’s federated design is its superpower. It allows the network to scale rapidly, connecting new data sources from public health labs, hospitals, and research centers without the massive bottlenecks and security reviews that hampered the early COVID-19 response. This agile approach means researchers can pivot to analyze new threats almost instantly, without waiting months for data transfers and legal agreements. The infrastructure is always ready.
NIAID’s Role in the NIH Data Revolution
The federated data ecosystem niaid is a cornerstone of the broader NIH Strategic Plan for Data Science. This strategy is being put into action by the NIH Cloud Platform Interoperability (NCPI) program, a groundbreaking effort to make the NIH’s largest and most important data platforms work together as a cohesive, federated whole.
The NCPI unites major NIH data initiatives, allowing researchers to perform cross-platform analysis. These platforms include:
- AnVIL: A cloud-based environment for genomic data analysis, focused on the vast datasets produced by the National Human Genome Research Institute (NHGRI).
- BioData Catalyst: A cloud ecosystem for heart, lung, blood, and sleep research, powered by the National Heart, Lung, and Blood Institute (NHLBI).
- Cancer Research Data Commons (CRDC): A comprehensive data ecosystem for the cancer research community, from the National Cancer Institute (NCI).
- Kids First Data Resource Center: A platform dedicated to fostering research into childhood cancer and structural birth defects.
- NCBI Resources: Foundational databases like dbGaP, which serves as a repository for data from studies that have investigated the interaction of genotype and phenotype.
What ties all these powerful platforms together? The FAIR Guiding Principles—a set of community-developed standards to make data Findable, Accessible, Interoperable, and Reusable. FAIR is the common language that allows these federated systems to communicate, ensuring that research is reproducible, collaborative, and efficient.
- Findable: Data is assigned a globally unique and persistent identifier (like a DOI for a paper). It is described with rich metadata that allows both humans and machines to discover it through search portals.
- Accessible: The protocols for accessing the data are standardized and open (e.g., via a specific API). This does not mean the data is open to everyone; it means the process for requesting and gaining access is clear and machine-readable, even when it requires authentication and authorization.
- Interoperable: The data uses common, shared vocabularies, ontologies, and formats. This allows a researcher to integrate and analyze datasets from different sources without extensive manual cleanup and harmonization.
- Reusable: The data has clear provenance (where it came from) and a license that specifies how it can be used. This gives researchers the confidence to build upon existing work.
Lessons from the earlier NIH Data Commons Pilot informed this mature shift toward federated interoperability. The pilot highlighted the limitations of centralization and underscored the need for an approach that keeps data secure at its source while enabling powerful collaborative analysis, fully respecting institutional autonomy.

The Road Ahead: Challenges and Opportunities
Building a federated ecosystem is a monumental technical achievement, but keeping it running, growing, and relevant is the real challenge. Our experience helping governments and public health agencies implement these systems highlights several key areas for long-term success.
- Sustaining Momentum and Governance: A federated system requires a robust governance model. This involves creating multi-institutional committees to set policies, define data access procedures, and resolve disputes. Continuous investment in maintenance, user support, and training is non-negotiable.
- Measuring Impact: To justify ongoing investment, the ecosystem’s leaders must define and track key performance indicators (KPIs). How much faster is research? How many new collaborations have been enabled? How many publications or discoveries have resulted? Demonstrating a clear return on investment is crucial.
- Harmonizing Data: While federation avoids moving data, semantic harmonization is still critical. For example, one hospital might code a patient’s sex as “M”/”F,” while another uses “1”/”2.” A federated query can’t work without a shared data model or on-the-fly translation using ontologies. This requires patient, ongoing work with data stewards.
- Setting and Enforcing Standards: The success of the ecosystem depends on universal adoption of standards like FAIR principles and common APIs. This requires patient consensus-building across diverse communities and strong incentives for compliance.
- Winning Hearts and Minds: The biggest challenge is often cultural, not technical. Researchers are accustomed to working with data they “own” on their local servers. The NDE must relentlessly prove its value by saving researchers time, enabling previously impossible science, and providing clear benefits that outweigh the learning curve. Our experience with Federated Data Governance has taught us that getting the human element right is the ultimate driver of adoption.
Despite these challenges, the future is incredibly promising. The federated data ecosystem niaid represents a fundamental and necessary shift in collaborative research, proving we can accelerate discovery without compromising security. This foundation could be the difference between a controlled local outbreak and a global crisis during the next health emergency.
FAQs: What You Need to Know About the NIAID Data Ecosystem
How do researchers get access to the NDE?
To get started, visit the NIAID Data Ecosystem Discovery Portal. This single interface lets you search all available datasets at once.
Crucially, data owners remain in control. After finding a dataset, you request access directly from the data steward. This ensures appropriate and secure use, as the data never leaves its source.
This federated approach combines easy findy with strong, localized security and control. Institutions don’t have to give up sovereignty to make their data findable.
What makes the NDE different from a centralized data commons?
The difference is fundamental. A centralized data commons copies all data to one location, creating security, compliance, and control issues.
The federated data ecosystem niaid does the opposite: data stays put, and analysis tools are brought to it. Researchers can query multiple sources, but the raw data never moves.
This is a game-changer for biomedical research, eliminating the legal and logistical nightmares of data transfer. It means faster setup and peace of mind for data owners.
How does the NDE make data from different sources work together?
How can scattered data be used together? The answer is smart standards. The NDE relies on the FAIR Guiding Principles (Findable, Accessible, Interoperable, Reusable) as its foundation.
In practice, each repository uses common metadata standards. This ‘universal language’ allows meaningful searches across different repositories.
The NDE also uses standardized APIs (Application Programming Interfaces) that allow analysis tools to query distributed data sources as if they were a single system.
The result is powerful cross-repository analysis without merging databases or restructuring data. This interoperability layer allows diverse datasets to work together while respecting institutional autonomy.
At Lifebit, we’ve perfected this harmonization approach, creating the connective tissue that open ups the full potential of valuable data.
Conclusion: Don’t Get Left Behind—Federation Is the Future of Biomedical Research
The federated data ecosystem niaid is more than a technical upgrade; it marks a new era of biomedical research where collaboration and speed don’t require sacrificing security or control.
NIAID’s $7.5M investment created an ecosystem where data stays put, accelerating research on COVID-19, AIDS, and other diseases. The NDE democratizes data access, preserving data sovereignty while shrinking research timelines from months to days.
This shift is happening across the NIH. The NCPI program is creating an interoperable network of platforms guided by FAIR principles, moving away from the flawed centralized model.
At Lifebit, we build the infrastructure that makes this possible. Our federated AI platform powers secure access to global biomedical data for biopharma and government partners. We’ve seen how Trusted Research Environments and federated governance transform research by bringing computation to the data.
The shift to federation is here. Organizations embracing it are accelerating research and cutting costs. Those who don’t risk being left behind, stuck with the outdated data silos federation solves.
Ready to see what federation can do for your research? Discover how federated platforms can transform your research and join us in building the infrastructure that will power the next generation of biomedical breakthroughs.

