Unlocking Genetic Secrets: AI’s Impact on Genomics

The Genomic Data Tsunami: Why AI Is No Longer Optional
AI for genomics is revolutionizing genetic data analysis, using machine learning to identify disease-causing variants, predict patient outcomes, and accelerate drug findy. These algorithms interpret the massive datasets from modern sequencing technologies, accomplishing tasks that would take humans years to complete manually.
Key applications of AI in genomics include:
- Variant calling and pathogenicity prediction – Identifying disease-causing mutations with higher accuracy.
- Drug target findy – Finding genes where variants protect against disease, revealing new therapeutic opportunities.
- Personalized medicine – Predicting individual disease risk and treatment response.
- Multi-omics integration – Combining genomic, transcriptomic, and proteomic data for a complete biological picture.
- Gene editing optimization – Improving CRISPR and other editing tools through deep learning.
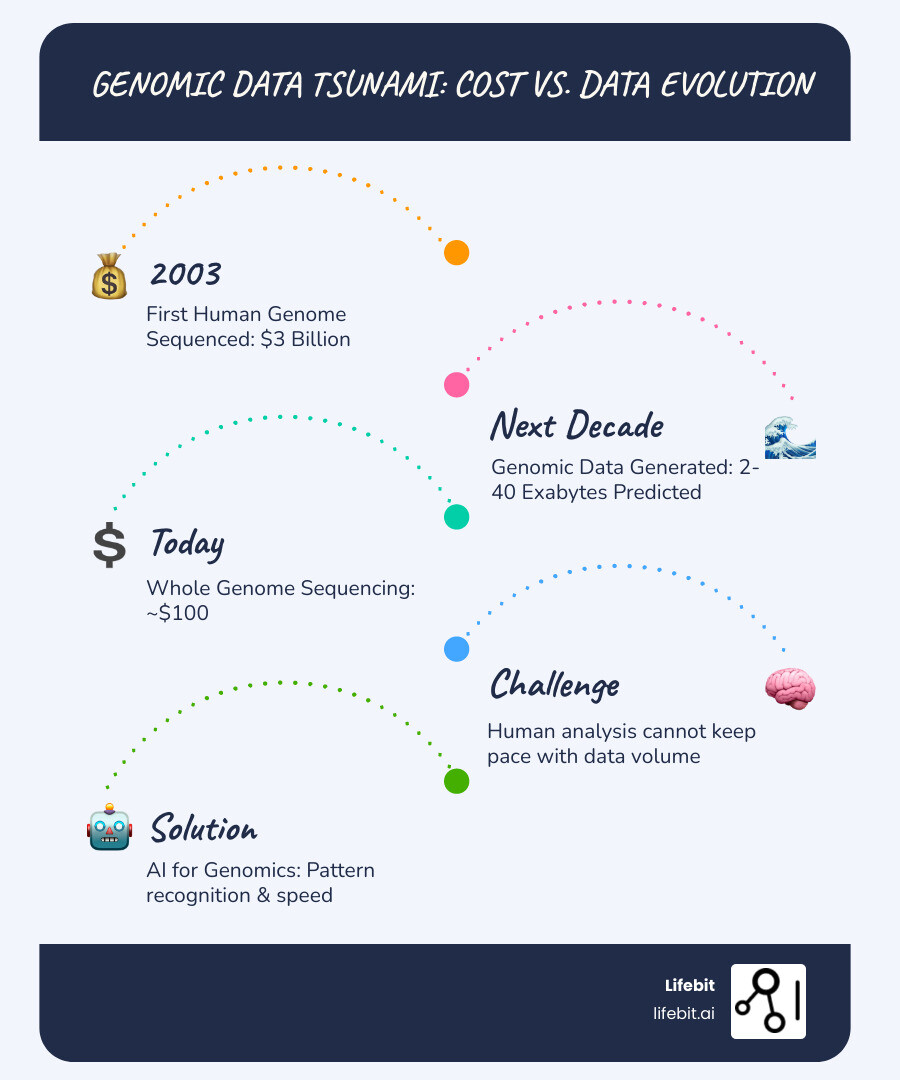
The numbers tell a striking story. The first human genome cost $3 billion to sequence in 2003. Today, whole genome sequencing can cost as little as $100—a 30-million-fold reduction. This has triggered an explosion of genomic data, with estimates predicting 2 to 40 exabytes generated in the next decade. For perspective, 40 exabytes is more storage than required for every word ever spoken in human history.
This data deluge has created an urgent problem: human analysis cannot keep pace. A single genome generates over 100 gigabytes of data, and traditional computational methods struggle with this volume. With the end of Moore’s Law, faster processors alone are not the answer.
This is where AI becomes essential. Machine learning excels at finding subtle patterns in massive, high-dimensional datasets. For example, GPU-accelerated AI has slashed germline variant calling from 16 hours to less than five minutes. The change extends beyond speed; AI models improve the accuracy of variant calling by learning from millions of examples. Generative AI is now designing novel proteins, and multi-omics platforms powered by AI are revealing biological insights that single data types miss entirely.
As CEO and Co-founder of Lifebit, I’ve spent over 15 years at the intersection of computational biology, AI, and genomics. The challenge of making AI for genomics accessible and actionable across secure, federated environments is what drives our work—enabling researchers to open up genetic insights without the barriers of data silos and computational bottlenecks.
This shift to AI-powered interpretation represents a fundamental change in how we understand biology, diagnose disease, and develop treatments. This guide explores how AI is reshaping every stage of genomic research.
What is AI and How Does it Apply to Genomics?
Artificial Intelligence (AI) is a collection of technologies designed to mimic or surpass human intelligence. Unlike traditional programming, AI systems learn from data. For a more in-depth exploration, we recommend A simple guide to key concepts in AI.
Two subfields are particularly impactful in genomics:
- Machine Learning (ML): Algorithms learn patterns from data without being explicitly programmed. In genomics, ML can identify genetic variations or predict disease susceptibility by training on vast amounts of labeled genomic data.
- Deep Learning (DL): A more advanced form of ML using artificial neural networks inspired by the human brain. These networks uncover incredibly complex patterns, such as interpreting raw signals from sequencing machines to infer DNA sequences (base calling) or predicting the pathogenicity of a genetic variant.
Why AI is the Perfect Match for Genomic Data
AI has become indispensable for genomics due to the nature of the data itself:
- Data Volume: Genomics generates data on an unprecedented scale. AI algorithms are built to process massive datasets efficiently.
- Data Complexity: Genomic data involves billions of base pairs and intricate interactions. AI, especially deep learning, excels at identifying non-obvious patterns within this high-dimensional data.
- High-Dimensionality: A single genome has millions of potential variations. AI models can handle this high number of features to make predictions.
- Speed of Analysis: Time is critical in clinical settings. AI dramatically reduces analysis time, turning weeks of work into minutes. This speed is game-changing for rapid diagnostics.
- Computational Power: The computational demand of genomics is immense. As traditional processor improvement slows, specialized hardware and accelerated computing are crucial for keeping up with the data tsunami. As we approach the end of Moore’s law, AI-driven computing is the only viable path forward.
Key Applications: How AI for Genomics is Accelerating Findy

The real power of AI for genomics is clear when you see it in action. Across research and clinical settings, AI is changing how we process raw sequencing data, call variants, predict disease, and optimize gene editing. What once took months now happens in hours—and with better accuracy.
Let’s look at where AI is making the biggest impact, from the technical backbone of genomic analysis like base calling and sequence alignment to the clinical applications that directly affect patient care.
Finding the Needle in the Haystack: AI-Powered Variant Calling
Imagine searching three billion letters of genetic code for the few typos that cause disease. That’s variant calling. Some variants are harmless; others are the difference between health and disease.
Traditional variant calling methods are slow and often miss subtle changes or flag false positives. This is where deep learning shines. Some algorithms treat variant calling like an image recognition problem, looking at aligned sequencing reads as if they were pictures. They learn to distinguish real variants from sequencing artifacts with remarkable accuracy. A prime example is Google’s DeepVariant, which reframed variant calling as an image classification task. It generates pileup images of sequence reads aligned around a potential variant and uses a convolutional neural network to classify the site. By training on validated genomes, DeepVariant achieved superior accuracy over previous methods, especially for challenging insertions and deletions (indels). This approach, which won multiple precisionFDA challenges, shows how techniques from other AI domains can create breakthroughs in genomics.
The results are dramatic improvements in accuracy for both germline variant calling (inherited changes) and somatic variants (acquired changes, like in cancer). These models catch single nucleotide variants, insertions, and deletions that older tools might miss.
Speed also matters, especially in clinical settings. GPU-accelerated systems have transformed analysis timelines—what took over 16 hours on traditional CPUs now runs in under five minutes. For a critically ill newborn with a suspected genetic disorder, that time difference can be lifesaving.
False positive reduction is another game-changer. Advanced deep learning tools can filter out noise from analysis results, meaning clinicians can focus on variants that truly matter instead of chasing down dozens of red herrings.
Predicting Disease and Optimizing Treatments
Finding variants is just the beginning. The harder question is: what do they mean? AI for genomics excels at answering this by learning from millions of previous cases.
Take genetic disorder identification—AI models can now analyze facial features to identify genetic disorders with surprising accuracy. This doesn’t replace genetic testing but helps flag which patients need deeper genomic investigation.
In cancer genomics, AI is transformative. Machine learning can predict cancer progression by analyzing tumor genomics alongside clinical data. It can identify the primary cancer type from a liquid biopsy—a simple blood test—allowing for earlier, less invasive detection.
Personalized medicine becomes real when AI predicts how your specific genetic makeup will respond to treatment. Will this chemotherapy work? Are you at risk for severe side effects? AI models trained on vast pharmacogenomic databases can answer these questions before treatment begins.
Even cutting-edge technologies like CRISPR get a major boost from AI. The success of CRISPR gene editing hinges on designing the perfect guide RNA (gRNA) to direct the enzyme to the correct DNA location. A poor gRNA can lead to low efficiency or dangerous ‘off-target’ edits. Deep learning models predict both on-target efficiency and off-target risk by analyzing sequence features of the gRNA and target DNA. By computationally screening thousands of potential gRNAs, AI helps researchers select the most effective and safest candidates before starting lab experiments, accelerating the development of more reliable gene therapies.
The Next Frontier: Generative AI and Multi-Omics Integration

The journey of AI for genomics is taking a fascinating turn. We’re moving beyond analyzing what exists in nature to creating new biological designs and connecting the dots across multiple layers of biological data—giving us a view of human health that was previously impossible.
How Generative AI is Revolutionizing Genomics
Generative AI has captured the world’s imagination, but its most profound impact may be in designing the molecules of life. If traditional AI learns to recognize patterns, generative AI learns to create new ones. In genomics, this means we can design entirely new proteins, each customized to perform a specific function.
A landmark achievement in this space is an AI tool that can predict the three-dimensional structure of almost any protein with remarkable accuracy. A protein’s shape determines its function, and knowing that shape is essential for designing drugs that interact with it. What once took years of lab work can now happen in hours.
The implications are profound. The Sanger Institute recently launched the world’s first Generative and Synthetic Genomics programme, aiming to engineer biology like we engineer electronics. Instead of just reading the genome, we’re learning to write new chapters.
Generative AI can design synthetic DNA sequences, predict how genetic changes will affect gene expression, and help us understand the consequences of altering a single nucleotide. This shift from observation to creation represents a move from a descriptive science to an engineering discipline.
Creating a Complete Picture with AI-Powered Multi-Omics
Human biology is not just about your genes. To understand health and disease, we need to look at the whole story. This is where multi-omics comes in—and where AI for genomics truly shines.
Your body operates across multiple biological levels: genomics (instructions), transcriptomics (genes being read), proteomics (proteins being built), metabolomics (chemical byproducts), and epigenomics (gene regulation). Each of these “omics” generates massive datasets. Trying to integrate them manually is like trying to conduct an orchestra where each musician is in a different room.
AI excels at this integration challenge. Machine learning algorithms can harmonize these different data types, finding connections that would be invisible if we examined each layer separately. For researchers diving deeper, this review of machine learning for multi-omics data offers valuable insights.
The power of this integrated approach shows up in real clinical applications. For example, some AI platforms combine multi-omics data to predict outcomes for pancreatic cancer patients. Other tools use interpretable machine learning to find hidden biological findings in high-dimensional multi-omics datasets, revealing how different biological layers interact.
At Lifebit, we’ve built our platform specifically to handle this complexity. Our Trusted Data Lakehouse harmonizes diverse data types across federated environments, while our AI and ML capabilities enable researchers to analyze multi-omics datasets at scale. This means you can integrate genomic, transcriptomic, and proteomic data securely, even when that data lives in different locations.
The holistic view that multi-omics provides changes everything. Instead of asking “What genes does this patient have?” we can now ask “How are those genes being expressed and interacting?” This systems biology approach gives us a complete picture of disease, helping us understand why two patients with the same genetic variant might respond differently to the same treatment.
From Code to Cure: AI’s Impact on Drug Findy and Personalized Medicine
The ultimate measure of AI for genomics is whether it improves human health. That’s exactly what’s happening in two critical areas: drug findy and personalized medicine.
Slashing Drug Findy Timelines
Traditional drug development is slow and expensive, taking 10-15 years and billions of dollars, with most candidates failing. AI is changing these odds.
The change starts with identifying novel targets. AI can mine millions of genetic sequences to find genes where specific variants protect against disease. The finding that certain variants in the PCSK9 gene protected against cardiovascular disease, for example, pointed researchers toward a completely new class of cholesterol-lowering drugs.
Some algorithms, trained on DNA from different species, can predict which genetic variants are likely to cause disease, dramatically improving our ability to identify promising drug targets. This helps teams deselect ineffective candidates early, reducing failure rates and saving years of wasted effort.
Predicting drug efficacy is another area where AI shines. Instead of testing thousands of compounds in the lab, AI models can simulate how potential drugs will interact with their biological targets. This computational screening saves enormous time and resources.
AI can also analyze the genomic profiles of diseases to identify opportunities for repurposing existing drugs, offering a much faster path to new treatments since these drugs have already cleared major safety problems.
Tailoring Treatments to Your DNA
The same drug can work brilliantly for one person and do nothing for another. This variation is largely written in our DNA. AI is finally making it possible to read that code and use it to guide treatment decisions.
Pharmacogenomics—the study of how genes affect drug response—generates the kind of complex data that AI handles best. By analyzing an individual’s genetic variants alongside clinical outcomes from thousands of others, AI can predict how that person will likely respond to a specific medication.
In precision oncology, this approach is already saving lives. AI analyzes a tumor’s specific mutations and matches them to targeted therapies. Instead of a one-size-fits-all approach, oncologists can select treatments customized to the molecular profile of each patient’s cancer.
Minimizing adverse effects is another crucial benefit. AI can flag genetic variants that cause dangerous drug reactions before a prescription is written, helping clinicians choose safer alternatives.
We’re moving beyond “precision medicine” toward truly personal medicine, which is especially important for complex diseases. The federated approach that platforms like Lifebit enable is crucial here. Personalized medicine requires access to diverse genomic datasets, but that data must remain secure. By bringing the analysis to the data, federated platforms make it possible to develop AI models that work across different populations while respecting privacy.
The journey from genetic code to the right treatment is getting shorter every day. That’s the promise of AI for genomics in action—not just faster research, but better outcomes for patients.
Navigating the Pitfalls: Challenges and the Future of AI in Genomics
As transformative as AI for genomics is, it comes with challenges that demand thoughtful solutions and careful ethical stewardship.
Overcoming the “Black Box” and Data Silos
Several technical obstacles stand in the way of realizing AI’s full potential in genomics.
Data quality is foundational. AI models are only as reliable as their training data. Genomic data can suffer from sequencing errors, batch effects, and inconsistent annotations. “Garbage in, garbage out” is the rule.
Then there’s the “black box” problem. Many powerful deep learning models can’t easily explain why they reached a conclusion. This is a major barrier to clinical adoption. Explainable AI (XAI) is crucial; we need systems that provide transparent reasoning that clinicians and patients can trust.
Data security and privacy are paramount. Genomic data is personal. Storing it in centralized locations creates privacy risks, while locking it in institutional silos hinders collaborative research.
Federated learning offers a powerful solution. Instead of moving sensitive data, the AI model travels to the data. It learns from each dataset in its secure environment and shares only the learned patterns, not the raw data. This preserves privacy while enabling large-scale collaboration. At Lifebit, our federated AI platform is built for this purpose: enabling secure access to global biomedical data without compromising privacy.
Data harmonization is another challenge, especially with multi-omics data from different labs using different protocols. Before AI can find patterns, these datasets must be translated into a common language. Our platform’s built-in harmonization capabilities address this, making diverse datasets analysis-ready.
Finally, algorithmic bias is a critical issue. If training data predominantly represents one population, the AI model will perform best for that group and potentially fail others. This is a matter of health equity. We must ensure models are trained on diverse, representative datasets.
The Ethical Landscape and Future of AI for Genomics
The ethical considerations are as profound as the technical challenges. We’re dealing with the fundamental building blocks of human life.
Key ethical considerations we must steer include data privacy and security, protecting genetic information from misuse; informed consent, ensuring people understand how their data will be used; algorithmic bias, preventing models from perpetuating health disparities; transparency and explainability, making AI decisions understandable; equity of access, ensuring benefits reach everyone; and data ownership, clarifying who controls genomic data.
Regulatory bodies like the FDA are developing guidance for AI in medical products, signaling that these tools are moving from research to clinical reality. The NIH’s Bridge to Artificial Intelligence (Bridge2AI) program exemplifies this focus on responsible AI, emphasizing ethical development and diverse data representation.
Looking ahead, the future of AI for genomics promises more sophisticated models, seamless data integration, and robust solutions for interpretability and privacy. The translation from research to clinical practice will accelerate, making personalized medicine a present-day reality.
At Lifebit, we’re building our platform with these considerations at the core. Our Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence & Analytics Layer) components deliver real-time insights and secure collaboration while maintaining the highest standards of data governance. The future of genomics isn’t just about what AI can do—it’s about ensuring it does so responsibly and equitably.
Frequently Asked Questions about AI in Genomics
How is AI trained for genomic analysis?
AI models are trained much like a person learns to recognize objects. We feed deep learning models vast collections of labeled genomic datasets where the outcome is already known. For instance, to build a model that identifies pathogenic variants, we show it countless examples of known disease-causing mutations alongside benign ones. The AI learns to recognize the distinct patterns that separate the two.
During this training, the model constantly adjusts its internal parameters to minimize prediction errors. The quality of this training depends heavily on diverse, well-annotated data. The more high-quality examples we provide—representing different populations and genetic contexts—the more accurate and reliable the AI becomes. This is why platforms that can access and harmonize data from multiple sources are so valuable.
What is the difference between AI in genomics and traditional bioinformatics?
Traditional bioinformatics has been the backbone of genomics for decades, relying on statistical methods and predefined algorithms programmed by humans. These approaches are powerful but can struggle with the complexity of modern genomic data.
AI for genomics, particularly machine learning, takes a different approach. Instead of following pre-programmed rules, AI models learn complex, non-linear patterns directly from the data itself. They can find relationships that humans might never think to program, often leading to more accurate predictions.
Think of it this way: traditional bioinformatics gives you a map with specific routes marked out. AI gives you a GPS that learns the best routes by analyzing real-time conditions—including paths that weren’t on the original map. The two work together: bioinformatics provides foundational tools, while AI adds a layer of intelligent pattern recognition on top.
Can AI predict my risk for all genetic diseases?
Not yet. AI has made remarkable progress in predicting risk for certain well-studied diseases. For monogenic diseases (caused by a single gene mutation, like cystic fibrosis), AI can be quite accurate. For polygenic diseases (influenced by multiple genes, like heart disease), AI is also showing promise by analyzing how many genetic variants work together.
However, AI models are only as good as their training data. For many rare or complex diseases, we simply don’t have enough examples to build reliable predictive models. Furthermore, your genetic code is just one part of the equation. Environmental factors, lifestyle, and diet all play significant roles. AI models trained purely on genomic data can’t capture these external influences.
While AI is making rapid strides, a complete predictive map for all genetic diseases remains a future goal. Today, AI helps identify risks for a growing number of conditions and guides researchers toward a better understanding of disease.
Conclusion: The Dawn of a New Era in Medicine
We are at a remarkable turning point. The convergence of AI and genomics is reimagining how we understand health, diagnose disease, and develop treatments.
AI for genomics is turning raw genetic data into actionable insights at unprecedented speed. What once took analysts years—identifying disease-causing variants or predicting patient outcomes—now happens in hours. We are moving from drowning in data to making life-saving findings from it.
The impact spans the entire biomedical landscape. Drug findy timelines are being compressed. Personalized medicine is becoming a clinical reality as AI interprets individual genetic profiles to guide treatment. And generative AI is taking us beyond reading the genome to engineering biology itself.
This powerful technology comes with real responsibilities. We must tackle challenges head-on—ensuring data quality, making AI decisions explainable, protecting patient privacy, and eliminating algorithmic bias. The ethical frameworks we build today will determine whether these advances benefit everyone.
At Lifebit, we are committed to this responsible path. Our federated AI platform enables researchers and clinicians to access and analyze global biomedical data securely, without compromising privacy. Through our Trusted Research Environment, Trusted Data Lakehouse, and R.E.A.L. layer, we make it possible to collaborate across borders while keeping sensitive genomic information protected.
The future of precision medicine is here, powered by the intelligent combination of human expertise and AI capability. We are excited to help open up genetic insights that will improve lives around the world.
Discover how a federated data platform can accelerate your research

