Nextflow’s Brain Boost: How AI is Revolutionizing Pipeline Development

Why AI for Nextflow is Changing Scientific Pipelines
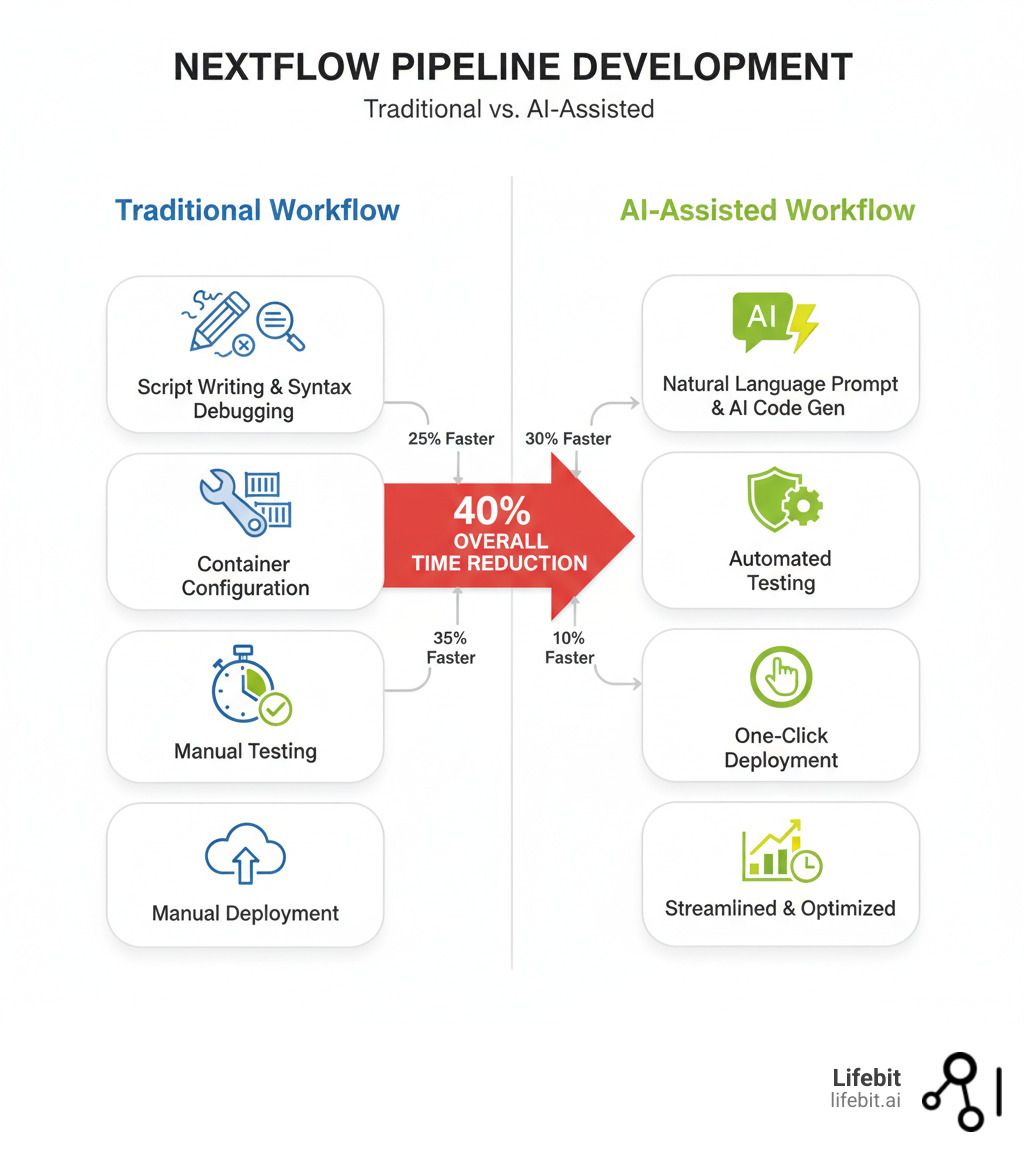
AI for nextflow is revolutionizing how scientists build computational workflows. It enables developers to generate production-ready pipelines from natural language, automatically debug complex errors, and convert legacy scripts to modern DSL2 syntax—reducing development time by up to 40%.
Quick Answer: What AI for Nextflow Can Do:
- Generate Code: Create complete, DSL2-compliant pipelines from simple descriptions.
- Debug Faster: Analyze execution logs and translate cryptic errors into actionable fixes.
- Modernize Scripts: Convert Bash, WDL, or CWL workflows to Nextflow in seconds.
- Automate Testing: Generate nf-test scripts and validate pipelines in AI-powered sandboxes.
- Ensure Best Practices: Produce modular, reproducible code that follows Nextflow standards.
The challenge in bioinformatics isn’t just writing code—it’s writing code that runs everywhere and scales effortlessly. Traditional pipeline development requires deep expertise in Nextflow, containerization, and cloud infrastructure. Even experienced developers spend hours debugging obscure errors or adapting scripts for different environments.
This is where AI changes everything. Instead of coding every process manually, scientists can describe their goals in plain English and let specialized AI assistants handle the heavy lifting. While general tools like ChatGPT are popular, they often produce outdated DSL1 syntax and lack the bioinformatics context for production-ready pipelines. Specialized AI for Nextflow, in contrast, understands genomic workflows, generates compliant DSL2 code, and integrates with testing frameworks.
For organizations managing siloed datasets, this AI-powered approach allows both expert and novice users to build and deploy compliant, scalable workflows without moving sensitive data, leading to faster insights and greater reproducibility.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit and a key contributor to Nextflow. I’ve seen how ai for nextflow is democratizing pipeline development, fundamentally changing who can build them and where they can run.
Why Nextflow is the Perfect Foundation for AI
If you’ve ever had a workflow break when moving from your laptop to an HPC cluster, you understand the barriers to reproducible science. Nextflow was designed to solve this, providing a language and runtime for orchestrating complex analyses that run anywhere, scale effortlessly, and produce identical results.
For over a decade, researchers have relied on Nextflow to orchestrate complex, data-intensive analyses. Its design principles—flexibility, parallelization, and portability—also make it the perfect foundation for ai for nextflow integration. The same structure that makes Nextflow reproducible also makes it predictable, which is exactly what AI needs to generate and optimize code effectively.
The Core Principles That Welcome AI Integration
Nextflow’s architecture provides a clear structure that AI can easily understand and work with.
- Unified parallelism: Its dataflow model abstracts the complexity of running tasks across thousands of nodes. AI can analyze the data dependencies between processes to suggest optimal parallel execution strategies, such as splitting large input files or adjusting the number of parallel tasks based on available compute resources, allowing AI to help optimize resource allocation without getting lost in the details of parallel computing.
- Stream-oriented processing: Data flows naturally from one step to the next. This reactive model is intuitive for AI to analyze, suggest dynamic optimizations, and easily trace data provenance, which is crucial for debugging and generating auditable workflows.
- DSL2 syntax: This modern syntax makes pipelines modular and readable. When AI generates DSL2 code, it’s following best practices that make workflows easier to maintain. AI models trained on DSL2 can also generate reusable modules and subworkflows, promoting a \”Don’t Repeat Yourself\” (DRY) approach that is a cornerstone of good software engineering.
- Modularity: Pipelines are broken into independent processes communicating through channels. This allows AI to refine individual components without breaking the entire workflow. For example, a developer can ask the AI to \”optimize the STAR alignment process,\” and it can focus solely on that module’s script, resource requirements, and container, without needing to re-evaluate the entire pipeline.
- Continuous checkpoints: Nextflow automatically tracks intermediate results, so a failed pipeline can be resumed from the last successful step. This resilience is invaluable for AI-assisted iteration, as the AI can leverage this feature by analyzing logs from a failed run to debug and fix the problematic step without re-running the entire workflow from scratch.
- Separation of logic and configuration: Core workflow functionality is kept separate from execution settings. This allows ai for nextflow tools to act as \”infrastructure experts,\” managing configurations for different environments (laptop, HPC, cloud) independently by generating different
profileblocks within anextflow.configfile.
These principles, highlighted in research like Nextflow enables reproducible computational workflows research, transform AI from a simple code generator into an intelligent partner that understands the structure and intent of your scientific workflows.
How Containerization Boosts AI-Generated Workflows
Nextflow’s deep integration with container technologies like Docker and Singularity solves the \”it worked on my machine\” problem. Containers package everything a pipeline needs—software, dependencies, and environment variables—into self-contained units that run identically everywhere.

This dramatically amplifies the power of AI-generated workflows. When a specialized AI creates Nextflow code, it can also suggest the optimal container images for the tools in your pipeline, effectively automating dependency management. For instance, if you ask the AI to \”use the latest stable version of Samtools,\” it can find the corresponding Biocontainers image (e.g., biocontainers/samtools:1.15--h3842212_0), ensuring the correct version is used and documented in the code. This eliminates manual searches on Docker Hub and reduces the risk of using incompatible tool versions.
Most importantly, containerization ensures code generated by AI runs reliably anywhere. The AI understands the nuances of different computing environments and writes portable pipelines, reducing manual tweaks and saving time. For organizations with federated environments, this combination is critical for deploying scalable workflows across multiple sites without moving sensitive data.
From Generalist to Genius: Choosing the Right AI for Nextflow
Not all AI is created equal, especially for building Nextflow pipelines. While 40% of developers use ChatGPT and 24% use GitHub Copilot for general coding, these tools often fall short for the specialized needs of ai for nextflow, leading to hours of debugging and refactoring.
The difference between a generalist and a specialized AI is whether your code works the first time, follows modern best practices, and scales reliably.
The Pitfalls of Using General AI for Nextflow Code
General AI models are trained on vast, diverse codebases, making them versatile but lacking deep domain expertise. For Nextflow, this leads to common problems:
- DSL1 vs. DSL2 Syntax Confusion: General AI often defaults to Nextflow’s outdated DSL1 syntax because its training data includes years of legacy code. For instance, it might generate a script where inputs are defined with
valand passed imperatively, instead of using DSL2’s declarativeinput:andoutput:blocks. This not only creates legacy code but also breaks the modularity that DSL2 is designed to provide, requiring significant manual refactoring. - Lack of Bioinformatics Context: These tools don’t understand the specifics of a genomic workflow. A user might ask for a variant calling pipeline, and a general AI could suggest using BWA for alignment followed by GATK HaplotypeCaller but fail to include the critical intermediate step of marking duplicates, leading to scientifically invalid results. They might suggest non-existent tools, incompatible software versions, or create pipelines that are scientifically nonsensical.
- Hallucinated and Inefficient Code: AI can generate plausible-looking but non-existent commands or parameters, such as inventing a command-line flag like
fastqc --auto-trim(which doesn’t exist). It may also produce inefficient code that ignores Nextflow best practices, such as using acollect()operator that aggregates all intermediate files into memory, causing a pipeline to crash on large datasets where a streaming approach would be necessary.
The Power of Specialized AI for Nextflow Development
Purpose-built ai for nextflow tools transform the development experience. Trained specifically on Nextflow DSL2, bioinformatics workflows, and community best practices, they offer significant advantages:
- Bioinformatics-Aware Models: They understand your scientific context. Ask for an RNA-Seq pipeline, and the AI knows the required steps, tools (FastQC, STAR, Salmon), and data formats (FASTQ, BAM).
- DSL2 as the Default: Specialized tools automatically generate code using modern DSL2 syntax, ensuring your pipelines are built on a solid, current foundation.
- Best Practice Enforcement: They produce modular code with clear separation of concerns, proper channel handling, and sensible resource management defaults, making pipelines easier to maintain and extend.
- Integrated Testing and Validation: These tools connect with frameworks like nf-test to help generate test scripts alongside your pipeline. Some even provide sandboxes to validate execution before deployment.
The Role of Curated Training Data
The ‘genius’ of specialized AI stems from its training data. These models are not just trained on a random scrape of GitHub; they are fine-tuned on high-quality, curated codebases like the nf-core community collection, which contains peer-reviewed, best-practice pipelines. This training ensures the AI learns from code that is not only functional but also robust, scalable, and adheres to community standards for documentation, structure, and testing. By learning from the best, the AI is predisposed to generate code of similar quality, effectively giving every developer an expert partner.

Choosing specialized AI for Nextflow means spending your time on research questions, not debugging syntax. It ensures what you build is robust, scalable, and ready to run anywhere.
A Practical Guide: 4 Steps to Build Pipelines with AI for Nextflow
The magic of ai for nextflow is how it transforms daily work. Instead of wrestling with syntax, you can have a conversation with an AI assistant that understands your goals. Here are four practical steps showing how this works.
Step 1: Generate Compliant DSL2 Code from a Simple Prompt
Instead of writing boilerplate code, you can simply describe your needs in plain English. A prompt like, \”Create a Nextflow DSL2 pipeline for RNA-Seq. It should take a samplesheet CSV as input. For each sample, it should run FastQC for quality control, then STAR for alignment against a reference genome, and finally Salmon for quantification. Use channels to pass data between processes,\” generates a complete, modular DSL2 workflow in moments.
The AI understands both Nextflow syntax and the bioinformatics context, ordering the steps correctly and defining the necessary processes and channels. For example, it might generate a STAR process that looks like this:
process ALIGN_STAR {
publishDir \"${params.outdir}/star\", mode: 'copy'
input:
tuple val(meta), path(reads)
path index
output:
tuple val(meta), path(\"*.bam\"), emit: bam
script:
\"\"\"
STAR --genomeDir ${index} \\
--readFilesIn ${reads[0]} ${reads[1]} \\
--runThreadN ${task.cpus} \\
--outSAMtype BAM SortedByCoordinate
\"\"\"
}
This natural language approach turns concepts into functional pipelines in minutes, freeing you to focus on the science.
Step 2: Modernize Legacy Scripts in Seconds
Old Bash, WDL, or CWL scripts that are difficult to scale can be modernized instantly. AI tools trained on multiple workflow languages can convert legacy code to modern Nextflow DSL2. Simply provide the old script and ask: \”Convert this Bash loop to a parallel Nextflow DSL2 process.\”
Before (Bash Script):
#!/bin/bash
SAMPLES=( \"sample1\" \"sample2\" \"sample3\" )
for SAMPLE in \"${SAMPLES[@]}\"; do
fastqc ${SAMPLE}_R1.fastq.gz ${SAMPLE}_R2.fastq.gz
done
After (AI-Generated Nextflow Process):
process FASTQC {
input:
tuple val(meta), path(reads)
output:
path \"*fastqc.html\"
script:
\"\"\"
fastqc ${reads[0]} ${reads[1]}
\"\"\"
}
The AI correctly identifies the looping pattern and converts it into a parallelizable Nextflow process that accepts data from an upstream channel. This preserves the scientific logic while making the workflow portable, scalable, and maintainable.
Step 3: Decode Errors with AI-Powered Log Analysis
Debugging cryptic Nextflow error logs is a common frustration. When a pipeline fails, you can copy the error log and ask the AI: \”What went wrong here and how do I fix it?\”
Cryptic Nextflow Error:
Caused by: java.lang.OutOfMemoryError: Java heap space -- Check script 'STAR' execution
AI-Powered Explanation:
*\”The error ‘java.lang.OutOfMemoryError’ means that the process named ‘STAR’ ran out of memory. This often happens during alignment with large reference genomes. The process was likely allocated the default amount of memory, but it required more.
Suggested Fix: Increase the memory allocation for this process in your nextflow.config file. Try setting memory = '32.GB' for the STAR process using a process selector like this:
process.withName('STAR').memory = '32.GB'
This will provide the task with more resources.\”
This transforms what could be hours of frustrating detective work into a quick, productive conversation.
Step 4: Automate Validation with AI-Generated Tests
Testing is critical for reproducibility but is often skipped due to its tedious nature. AI for nextflow automates the creation of test scripts for frameworks like nf-test.
You can ask the AI to \”Generate an nf-test script for my FastQC process that checks if the output zip and html files are created correctly.\” The AI will produce a complete test definition with assertions.
AI-Generated nf-test Script:
test(\"fastqc process should produce a zip and html file\") {
when {
process {
"""
input[0] = [ [id:'test'], [ file(params.test_data_r1), file(params.test_data_r2) ] ]
"""
}
} then {
assert process.success
assert snapshot(process.out).match()
}
}
Specialized tools can even create a sandboxed micro-VM with Nextflow pre-installed to validate the code safely. This integrates testing into the development workflow, ensuring pipelines are not just functional but genuinely reliable before deployment.
The Bigger Picture: How AI Accelerates Scientific Findy
The combination of AI and Nextflow is more than a coding accelerator; it’s changing how science is conducted, who can participate, and how quickly findies are made.
Enhancing Reproducibility and Portability at Scale
The “works on my machine” problem has long plagued computational biology. AI for nextflow addresses this by generating code that is inherently portable. The AI not only writes workflow logic but also suggests the correct Docker or Singularity container, ensuring the entire software environment is packaged with the pipeline. This guarantees that the code runs identically on a laptop, HPC cluster, or in the cloud.

Furthermore, AI can generate optimized configuration files for different platforms, as detailed in the Nextflow documentation on executors. It can create configs for Kubernetes, SLURM, or AWS Batch that handle resource allocation and cost management automatically. This makes advanced infrastructure accessible without deep expertise and ensures true scientific reproducibility at scale.
Democratizing Science for a New Wave of Researchers
AI for nextflow is breaking down the technical barriers that have limited computational biology to programming experts. Biologists and clinicians can now translate their scientific questions into functional pipelines by describing their needs in plain English. This lowering of the barrier to entry brings more diverse perspectives into computational research.
This democratization enables new research paradigms. At Lifebit, our federated AI platform combines AI-powered Nextflow workflows with secure, distributed data access. Researchers can run complex analyses on datasets from multiple institutions without moving sensitive patient information. This empowers pharmaceutical companies to analyze real-world evidence from hospitals globally while maintaining strict privacy compliance.
This empowerment of non-programmers accelerates research cycles. When the time from question to analysis shrinks from weeks to hours, scientists can iterate faster and test more hypotheses. This is especially critical for secure analysis on distributed data, a core capability for fields like precision medicine. With Lifebit’s Trusted Research Environment and AI-powered workflows, global collaboration becomes possible while upholding the highest standards of data governance. The technical barrier is dissolving, accelerating not just pipelines, but the pace of scientific findy itself.
Frequently Asked Questions about AI and Nextflow
Can AI write a complete, production-ready Nextflow pipeline?
Yes, specialized AI tools can. Unlike generalist AI, they are trained on Nextflow’s DSL2 syntax, bioinformatics workflows, and community best practices. They can take a simple prompt and generate a complete, functional, and modular pipeline with all necessary configurations, process definitions, and even parameter schemas (nextflow_schema.json). This can cut development time by up to 40%, allowing researchers to focus on science instead of syntax.
How does AI help analyze complex Nextflow execution logs?
AI acts as an intelligent debugging partner. When a pipeline fails, you can provide the error log to an AI assistant and ask what went wrong. The AI parses the cryptic output, identifies the root cause (e.g., a memory issue, incorrect file path, or container error), explains it in plain English, and suggests a specific, actionable fix. This reduces troubleshooting time from hours to minutes, especially for those new to Nextflow.
What are the best AI tools for generating Nextflow code?
While general tools like ChatGPT are popular, the most effective tools are specialized AI assistants purpose-built for the Nextflow ecosystem. These are often integrated into IDEs like VS Code and provide context-aware assistance. They understand DSL2 as the standard, recognize bioinformatics tools, and enforce best practices for modularity, containerization, and testing. This ensures the generated code is not just functional but also robust, scalable, and reproducible.
How does AI for Nextflow handle security and data privacy?
This is a critical consideration, especially in biomedical research. Leading ai for nextflow tools address this in two ways. First, the AI assistant itself often runs within a secure environment, such as a VS Code extension on a local machine or within a private cloud, meaning your code and prompts do not need to be sent to a public third-party service. Second, the AI is designed to generate code that works within secure data environments. For example, it can generate pipelines that are fully compatible with Trusted Research Environments (TREs). The AI doesn’t access the sensitive data itself; it simply writes the workflow logic that will be executed securely within the TRE, where the data resides. This separation of code generation from data execution ensures that intellectual property and patient data remain protected.
Conclusion: The Future is Collaborative and AI-Powered
The integration of ai for nextflow is fundamentally reshaping scientific research. As we’ve seen, Nextflow’s reproducible and portable design provides the perfect foundation for AI. Specialized AI assistants, unlike generalist tools, understand the nuances of bioinformatics, generating compliant DSL2 code, modernizing legacy scripts, decoding errors, and automating tests—slashing development time by up to 40%.
This isn’t just about efficiency; it’s about accessibility. The barrier to entry for bioinformatics is crumbling, empowering researchers without coding expertise to build production-ready pipelines. This democratization accelerates the entire research cycle, from hypothesis to findy.
At Lifebit, we’ve built our federated AI platform on this vision. We bring computational power directly to the data, enabling researchers to run sophisticated ai for nextflow pipelines on sensitive, distributed datasets without moving them. Our platform, including the Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence & Analytics Layer), facilitates secure, large-scale research and collaboration across institutions and borders.
The shift from manual coding to AI-assisted design is here. It’s enabling faster, more reliable, and more accessible science. From AI-designed drug compounds to real-time safety surveillance, the impact is already tangible.
We believe in making accelerated scientific findy possible for everyone. The future of scientific computing is collaborative, AI-powered, and federated—and it’s changing biomedical science today.
Visit our platform to see how we’re making this vision real: https://lifebit.ai/platform/

