Don’t Panic: The Stress-Free Nextflow Setup Guide

Why Getting Your Nextflow Install Right Matters More Than You Think
Nextflow install is the first step to open uping scalable, reproducible computational pipelines for genomics and biomedical research. Here’s how to get it done fast:
Quick Install (Linux/macOS):
- Install Java 17+ – Download from Oracle or use SDKMAN
- Run install command –
curl -s https://get.nextflow.io | bash - Make executable –
chmod +x nextflow - Add to PATH – Move to
/usr/local/binor$HOME/.local/bin - Verify – Run
nextflow info
For Windows: Use WSL2 with Ubuntu 20.04 (see detailed steps below)
For Cloud: Skip local setup entirely with Lifebit Cloud
If you’re running federated AI research, working with siloed EHR data, or building multi-omic pipelines across secure environments, a broken Nextflow setup can kill your timeline before you even start.
Most install guides assume you’re working on a clean system with perfect permissions and no firewall rules. But in reality? You’re juggling Java versions, PATH variables, Docker conflicts, and Windows compatibility issues—all while your team waits for results.
The good news: Nextflow install doesn’t have to be painful. Whether you’re setting up locally on Linux, macOS, or Windows (via WSL2), or skipping the hassle entirely with Lifebit Cloud, this guide walks you through every step with zero fluff.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, and a key contributor to Nextflow itself. Over 15 years, I’ve helped thousands of researchers get their nextflow install right the first time—so they can focus on science, not system errors. Let’s get you running.
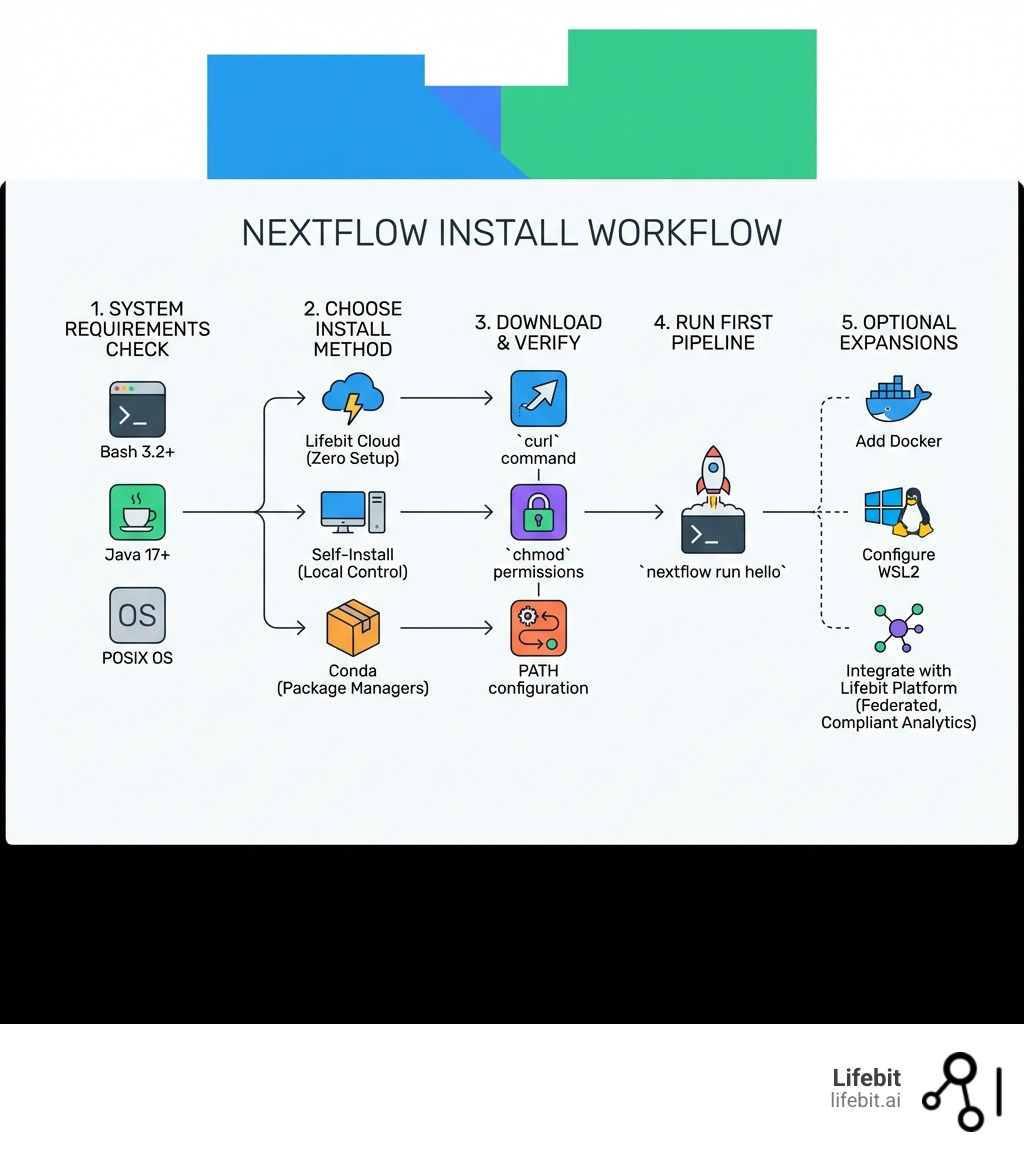
System Requirements: What You Need Before You Install Nextflow
Before we dive into the commands, we need to ensure your “engine room” is ready. Nextflow is built on the JVM (Java Virtual Machine), which makes it incredibly portable, but it does have specific tastes when it comes to versions. Understanding these requirements is crucial because a mismatch often leads to cryptic stack traces later in your pipeline execution.
To perform a successful nextflow install, your system must meet these core requirements:
- Bash 3.2 or later: This is the standard shell for most modern Linux and macOS systems. If you are on a legacy enterprise system, verify this with
bash --version. - Java 17 or later (up to 25): This is the most critical requirement. As of version 24.11.0-edge, support for Java versions prior to 17 has been dropped. This shift was necessary to leverage modern JVM features like enhanced garbage collection (G1GC) and better memory management, which are vital when handling high-throughput genomic data.
- POSIX Compatibility: Nextflow thrives on Linux, macOS, and Windows Subsystem for Linux (WSL). It relies on standard Unix-like file system behaviors for task execution and file locking.
At Lifebit, we often work with massive datasets where stability is non-negotiable. We recommend using a Long-Term Support (LTS) version of Java to avoid unexpected runtime glitches. While Nextflow can run on various JDK distributions, we have found the most consistent performance with Eclipse Temurin or Amazon Corretto.
| Feature | Lifebit Cloud Install | Local Self-Install |
|---|---|---|
| Setup Time | < 1 Minute | 10-20 Minutes |
| Java Dependency | Managed by Lifebit | Manual Install Required |
| OS Compatibility | Browser-based (Any OS) | Linux, macOS, WSL |
| Maintenance | Automatic Updates | Manual (self-update) |
| Scalability | Instant Cloud Bursting | Limited to Local Hardware |
Choosing and Managing Your Java Version
If your java -version returns anything lower than 17, or if the command isn’t found, don’t panic. The community consensus—and our own best practice at Lifebit—is to use SDKMAN! to manage your Java environments. SDKMAN! is a tool for managing parallel versions of multiple Software Development Kits on most Unix-based systems. It allows you to switch between versions without breaking your system’s default settings or requiring root access.
To get started with SDKMAN!, run:
curl -s "https://get.sdkman.io" | bash
source "$HOME/.sdkman/bin/sdkman-init.sh"
sdk install java 17.0.10-tem
Why Java 17? Beyond being the new baseline for Nextflow, Java 17 introduced significant performance improvements for the Groovy language (which Nextflow is written in). It handles large-scale object allocation more efficiently, which translates to a more responsive Nextflow head node when you are orchestrating thousands of concurrent jobs. For more detailed environment configurations, you can always refer to Lifebit’s Java best practices.
The Fastest Way to Install Nextflow
Once Java is ready, the actual nextflow install is a single-line affair. Nextflow is distributed as a self-installing “capsule”—a lightweight JAR file that contains the logic to download the rest of the framework upon its first execution. This ensures that the core binary remains small while still being able to pull in the full power of the Nextflow runtime.
Open your terminal and run:
curl -s https://get.nextflow.io | bash
If you prefer wget, this works too:
wget -qO- https://get.nextflow.io | bash
This creates a nextflow executable file in your current directory. To make it accessible from anywhere on your system, you need to change its permissions and move it to a folder in your $PATH. We recommend $HOME/.local/bin for user-level installs to avoid needing sudo permissions, which is often restricted on shared HPC clusters:
chmod +x nextflow
mv nextflow $HOME/.local/bin/
(Note: Ensure $HOME/.local/bin is in your PATH by adding export PATH=$PATH:$HOME/.local/bin to your .bashrc or .zshrc file. After adding it, run source ~/.bashrc to refresh your session.)
For a deeper dive into enterprise-grade deployment, check out Lifebit’s Nextflow install documentation.
Three Ways to Install Nextflow on Linux and macOS
While the curl method is the most popular, it’s not the only way. Depending on your environment—whether it’s a personal laptop, a locked-down HPC (High-Performance Computing) cluster, or a CI/CD runner—you might choose a different path. Each method has trade-offs regarding dependency management and version control.
- Self-Install (Recommended): The method described above. It’s fast, easy to update, and puts you in control. It downloads the
nextflowlauncher, which then manages the download of the specific framework version required for your pipelines. This is the most “Nextflow-native” way to work. - Conda/Bioconda: You can install via
conda install -c bioconda nextflow. This is popular in bioinformatics because it bundles the Java dependency. However, be warned: Conda packages can sometimes lag behind the official releases. Furthermore, Conda’s dependency solver can sometimes struggle with the specific version of Java required by newer Nextflow releases, leading to “environment solver” hangs. - Standalone Distribution: Ideal for offline or air-gapped environments. You can download the full package (often labeled as
nextflow-all) from the GitHub releases page. This version includes all runtime dependencies in a single large JAR file, meaning it won’t try to reach out to the internet to download the framework on its first run. This is essential for secure government or clinical environments where outbound internet access is strictly controlled.
For those running complex multi-omic workflows, we explore these methods in detail in our guide on Advanced Analytics with Nextflow Pipelines.
Using Lifebit Cloud for Your Nextflow Install
Let’s be honest: sometimes you just want to run a pipeline without fighting with your OS, Java versions, or PATH variables. This is where the Lifebit platform shines. When you use Lifebit, you get a fully managed Nextflow environment right out of the box, pre-configured for the most demanding biomedical workloads.
- Zero Conflicts: No more Java version “hell.” We maintain the optimal JDK for every Nextflow version.
- Instant Scaling: Local installs are limited by your CPU and RAM. Lifebit allows you to go from a single process to thousands of cloud nodes across AWS, Azure, or Google Cloud without changing a single line of your Nextflow code.
- Compliance Built-in: Ideal for sensitive biomedical data that cannot leave its home region. Lifebit brings the Nextflow installation to the data, rather than moving the data to the compute.
By using Lifebit Cloud, you bypass the local nextflow install problems entirely. Our platform handles the orchestration, so you can launch NF-core Nextflow Pipelines with a single click, backed by enterprise-grade security.
Integrating Containers for Reproducibility
A nextflow install is only half the battle. To ensure your science is reproducible, you shouldn’t install bioinformatics tools (like BLAST, Samtools, or GATK) directly on your machine. This leads to “it works on my machine” syndrome. Instead, use containers.
Nextflow has first-class support for container engines, allowing you to define the environment for every single task in your pipeline. This is managed via the process.container directive in your config file.
- Docker: The industry standard for local and cloud development. It provides excellent isolation but requires root or “docker group” privileges.
- Singularity/Apptainer: The go-to for HPC environments. It allows you to run containers as a standard user, which is a requirement for most shared university or institutional clusters.
- Conda: Useful for environments where container runtimes aren’t available. Nextflow can create and manage Conda environments on the fly for each process.
We strongly recommend Docker for local testing. Once installed, you can run any Nextflow pipeline with the -with-docker flag. Nextflow will automatically pull the necessary images from Docker Hub or Quay.io, mount the required volumes, and handle the cleanup. For more on this, see our Lifebit AI Nextflow Complete Guide.
Running Nextflow on Windows: The Lifebit WSL2 Power Setup
For a long time, Windows users were the “underdogs” of the Nextflow world, often forced to use slow Virtual Machines or limited Cygwin environments. That changed with the Windows Subsystem for Linux (WSL2), which provides a genuine Linux kernel inside Windows.
To get Nextflow running on Windows 10 or 11, follow these high-level steps to ensure a high-performance environment:
- Enable WSL2: Open PowerShell as Administrator and run
wsl --install. This will install the Linux kernel and the default Ubuntu distribution. - Install Ubuntu: We recommend the Ubuntu 22.04 LTS distribution from the Microsoft Store for the best compatibility with modern Java 17+ packages.
- Update Linux: Inside your new Ubuntu terminal, run
sudo apt update && sudo apt upgrade. This ensures your package manager is ready to install the necessary dependencies for Java.
Setting up a Windows environment can be tricky, especially when dealing with Trusted Research Environments (TREs) where security policies might block WSL2 networking. We’ve documented the specific problems in our post on Nextflow Cloud Challenges TRE Start.
Configuring VS Code and Git for Windows Development
To turn your Windows machine into a professional development station, you need the right IDE. Visual Studio Code (VS Code) is our top pick because of its deep integration with WSL2.
- Remote WSL Extension: This is the “magic” component. It allows VS Code to run its server component inside your Ubuntu instance while the UI runs on Windows. You get the speed of Linux file systems with the familiar UI of Windows.
- Nextflow Language Support: Install the Nextflow extension by nf-core. It provides syntax highlighting, code completion, and linting, which are essential for writing bug-free DSL2 code.
- Git: Essential for version control. Install it inside your WSL instance (
sudo apt install git) to keep your pipeline code synced with GitHub. Avoid using the Windows version of Git for files stored inside the WSL file system to prevent line-ending issues (CRLF vs LF).
If you’re looking to apply these tools to real-world tasks, check out our Nextflow DeepVariant Tutorial.
Optimizing WSL2 Performance for Nextflow
One common issue with Nextflow on WSL2 is memory consumption. By default, WSL2 can consume up to 80% of your total Windows RAM, which can lead to the host OS becoming sluggish. You can manage this by creating a .wslconfig file in your Windows user profile folder (C:\Users\):
[wsl2]
memory=8GB # Limits VM memory in WSL2
processors=4 # Limits the number of CPUs
kernelCommandLine = vsyscall=emulate
Furthermore, Docker Desktop now includes a WSL2 backend. This means Docker containers run natively on your Linux kernel rather than in a heavy Hyper-V VM. This provides massive performance gains, especially for I/O-heavy bioinformatics tasks. Always store your project files within the Linux file system (e.g., /home/user/projects) rather than the Windows mount (/mnt/c/), as the cross-OS file system performance is significantly slower.
For more troubleshooting on complex setups like Jupyter and RStudio integration within a Nextflow ecosystem, see Nextflow Challenges Jupyter RStudio.
Verifying and Updating Your Nextflow Environment
Once you’ve completed your nextflow install, you need to make sure everything is ticking correctly. Verification is a two-step process: checking the environment and running a test workload.
Run the following command to see your system’s heartbeat:
nextflow info
This command is your best friend for debugging. It will display your current Nextflow version, the system architecture (e.g., x86_64), the Groovy version, and the specific Java runtime details. If you see a version number and no errors, your installation is successful.
To test a real (but tiny) pipeline, run:
nextflow run hello
This command pulls a basic script from the Nextflow GitHub repository. If you see “Hello world!” printed in multiple languages, it confirms that Nextflow can successfully download remote scripts, compile them on the fly, and execute them on your local JVM.
Managing and Updating Nextflow
Nextflow moves fast. The community is incredibly active, with typically two stable releases a year and monthly “edge” releases that introduce cutting-edge features like improved AWS Batch integration or new DSL2 syntax.
- Self-update: To stay current, run
nextflow self-update. This will check for the latest stable version and replace your local binary. -
Specific Versions: In science, version stability is key. If you need an older version to reproduce a result from a 2022 paper, you don’t need to reinstall. Use the
NXF_VERenvironment variable:NXF_VER=23.10.1 nextflow run my-pipelineNextflow will automatically download the 23.10.1 runtime in the background and use it for that specific execution.
- Edge Releases: Want to try the newest features before they hit the stable branch? Set
export NXF_EDGE=1before running the update command.
Troubleshooting Common Installation Errors
Even with the best guide, things can go sideways. Here are the “Big Three” errors we see at Lifebit and how to fix them:
- “Java not found”: This usually means Java is installed but not in your system’s PATH. If you used SDKMAN!, ensure you have
source "$HOME/.sdkman/bin/sdkman-init.sh"in your.bashrc. You can verify your Java path withwhich java. - “Permission denied”: This happens if you forget
chmod +x nextflow. Without the executable bit set, Linux treats the file as a plain text file rather than a program. - “Unsupported class version error”: This is the classic “Java is too old” error. It means you are trying to run a modern Nextflow version (which requires Java 17) on an older Java 8 or 11 runtime. Update your Java version using SDKMAN! as shown in Section 1.
- Connection Timeouts: If you are behind a corporate firewall,
curlmight fail. You may need to set your proxy variables (export http_proxy=...) or download the standalone distribution manually.
If you’re still stuck, the Lifebit support team and the Nextflow Slack community are great places to get expert help. Most issues are related to environment variables, so always include the output of nextflow info and echo $PATH when asking for help.
Conclusion
A successful nextflow install is the foundation of modern, scalable research. By following this guide, you’ve moved past the most common roadblocks—Java versioning, PATH configuration, and OS compatibility—and are ready to build pipelines that can change the world. Whether you’re analyzing rare disease variants, processing vast multi-omic datasets, or building federated AI models, your environment is now robust and ready.
At Lifebit, we are dedicated to making this journey as smooth as possible. We understand that for many researchers, the goal isn’t to become a systems administrator, but to get to the science. Our platform doesn’t just run Nextflow; it improves it with federated AI capabilities, secure governance, and real-time analytics that work across any cloud or on-premise infrastructure.
Ready to take your pipelines to the next level? Accelerate Nextflow with Lifebit’s NF-Copilot Feature and see how we turn complex data into actionable insights in record time.
Frequently Asked Questions
How do I install a specific version of Nextflow?
You don’t need to reinstall or manually download files. Simply set the NXF_VER variable before your command, e.g., NXF_VER=22.10.0 nextflow run .... Nextflow is designed to be multi-version aware and will download the required runtime on the fly and store it in your ~/.nextflow directory.
Can I run Nextflow without a local installation?
Yes. You have two main options. First, you can use GitHub Codespaces, which provides a cloud-based terminal environment where you can install Nextflow temporarily. Second, and more powerfully, you can use the Lifebit Platform. Lifebit provides a full-featured, enterprise-grade experience that requires zero local setup, allowing you to manage data and pipelines through a secure web interface.
How does Lifebit support high-compliance research?
Lifebit focuses on federated AI and secure access to sensitive biomedical data (like EHRs and genomic biobanks). In many cases, this data cannot be moved to a central cloud due to privacy laws (like GDPR or HIPAA). Lifebit provides a Trusted Research Environment (TRE) that allows you to run Nextflow pipelines locally where the data resides, while still providing a unified cloud-like management experience.
Is Windows really viable for Nextflow development?
Absolutely. With WSL2 and the VS Code Remote extension, the experience is nearly identical to native Linux. You get the full power of the Linux command line and container support (via Docker Desktop) while maintaining your Windows workflow for email, documentation, and other tasks. It is a highly productive environment for modern bioinformaticians.
What is the difference between the nextflow binary and the .nextflow folder?
The nextflow binary you download is just a launcher. When you run it, it creates a hidden .nextflow directory in your home folder. This directory contains the actual framework JAR files, plugins, and the history of your pipeline runs. If you ever want to “reset” your Nextflow installation, you can safely delete this folder, and Nextflow will recreate it the next time it runs.
For more information or to join our growing community, visit the Lifebit Nextflow Community.

