Generative AI and OMOP: Revolutionizing Real-World Evidence

Stop Waiting Months for OMOP: How AI Turns Siloed RWD into Same-Week Insight
AI for OMOP is changing how healthcare organizations harmonize real-world data, slashing harmonization time from months to minutes while opening up insights from unstructured clinical notes, medical images, and multimodal sources that traditional methods miss.
Quick Answer: What AI Brings to OMOP
- Speed: Map data to OMOP in hours instead of months
- Completeness: Process unstructured clinical notes and multimodal data automatically
- Accuracy: AI-powered terminology mapping reduces errors and information loss
- Cost: Reduce ETL project costs by automating manual workflows
- Scale: Handle millions of patient records across distributed networks
For pharmaceutical leaders, regulatory agencies, and public health organizations, the pain is familiar: valuable insights are trapped in siloed, messy data. Electronic health records (EHRs), claims databases, and lab results all speak different languages. The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) aims to solve this by standardizing Real-World Data (RWD) into a unified format for cross-institutional research.
The catch? Converting raw data to OMOP is brutally slow. Traditional Extract, Transform, Load (ETL) processes take months, require specialized teams, and lose critical information, especially from unstructured sources like physician notes, which hold a large share of clinical context. By the time data is ready, the insights are stale.
This is where AI changes everything. Large Language Models (LLMs) and agentic AI workflows can now process structured EHR data, FHIR resources, claims records, and unstructured clinical notes in days or even hours. AI doesn’t just accelerate the old process; it transforms what’s possible, creating a more clinically complete patient data model for better pharmacovigilance, cohort finding, and real-world evidence generation.
The OHDSI Community has built powerful open-source tools for OMOP, but the manual burden remains high. AI-powered platforms like Lifebit’s are now emerging that automate data harmonization end-to-end, enabling federated analytics without moving sensitive data and empowering researchers to ask questions in natural language instead of writing complex SQL queries.
I’m Maria Chatzou Dunford, CEO of Lifebit. We build an AI-powered, federated platform to harmonize genomic, clinical, and real-world data in secure, distributed environments. AI for OMOP is core to our mission. In this guide, I’ll show you how AI is revolutionizing OMOP data change and what it means for generating real-time, research-ready insights.
Simple guide to AI for OMOP terms:
- AI for data harmonization
- FDA Sentinel 3.0
- AI for Federation
The OMOP Bottleneck: Why Manual Data Harmonization Costs You Time and Insights
The OMOP Common Data Model promises to open up insights from diverse healthcare data, but the journey to compliance is a major bottleneck for real-world evidence generation. Manual ETL processes are slow, costly, and error-prone, with projects stretching for months. This leads to information loss and inconsistent terminology mapping, especially with the vast amounts of unstructured data in modern clinical records.
A critical issue is the failure of traditional methods to process unstructured data. Clinical notes, physician narratives, and medical images hold invaluable patient information that is inaccessible to standard ETL pipelines. This creates data quality issues and an incomplete patient picture, hindering the accuracy and depth of downstream analyses.
Our work with biopharma, governments, and public health agencies highlights these primary CDM implementation challenges:
- Time and resource intensity: Converting diverse source data into a standardized format like OMOP requires significant manual effort, which can take months and delay critical research.
- Difficulty with unstructured data: A vast amount of clinical information resides in free-text notes. Traditional ETL struggles to extract, normalize, and map this data to OMOP concepts, leading to substantial information loss.
- Vocabulary and mapping inconsistencies: Variations in local coding practices and the sheer volume of concepts make consistent mapping a continuous challenge. Manual crosswalks are time-consuming to create and maintain.
- Data quality validation: Ensuring data accuracy and completeness after conversion is crucial, but manual validation is slow and cannot scale with modern healthcare datasets.
These challenges mean valuable insights remain locked away, leaving the full potential of RWD for improving patient care and accelerating medical findings untapped. Compared to traditional methods, AI solutions promise to dramatically reduce the time and effort involved.
The Struggle with Diverse Data Formats
The healthcare landscape is a mosaic of disparate data formats. Structured EHR data varies significantly between institutions. FHIR standards aim to improve interoperability but are not universally implemented. Claims data provides rich administrative information but lacks clinical depth.
The real treasure trove, and biggest headache, is unstructured clinical notes and multimodal data. Scanned documents, physician free-text notes, and medical images contain a large portion of a patient’s story. Traditional, rule-based ETL processes struggle to parse, understand, and map this data to a structured model like OMOP. This creates pervasive data silos and significant interoperability gaps, making it nearly impossible to build a comprehensive patient view.
Limitations of Traditional CDM Approaches
Traditional approaches to implementing CDMs like OMOP have inherent limitations. They often impose rigid structures that, while standardizing, can lead to a loss of data granularity. Important nuances in the original source data may be simplified during the conversion process, impacting the richness of subsequent analyses.
For distributed data networks, traditional CDM approaches are particularly inefficient. Each institution’s manual ETL process is a massive undertaking, which can create inconsistencies in mapping quality and makes study reproducibility a significant challenge. As one scientific review highlights, comparing different CDMs reveals implications for active drug safety surveillance, underscoring the need for more efficient and robust methods scientific research on CDM challenges. The effort required to maintain manual mappings means the “data ready” state is often fleeting.
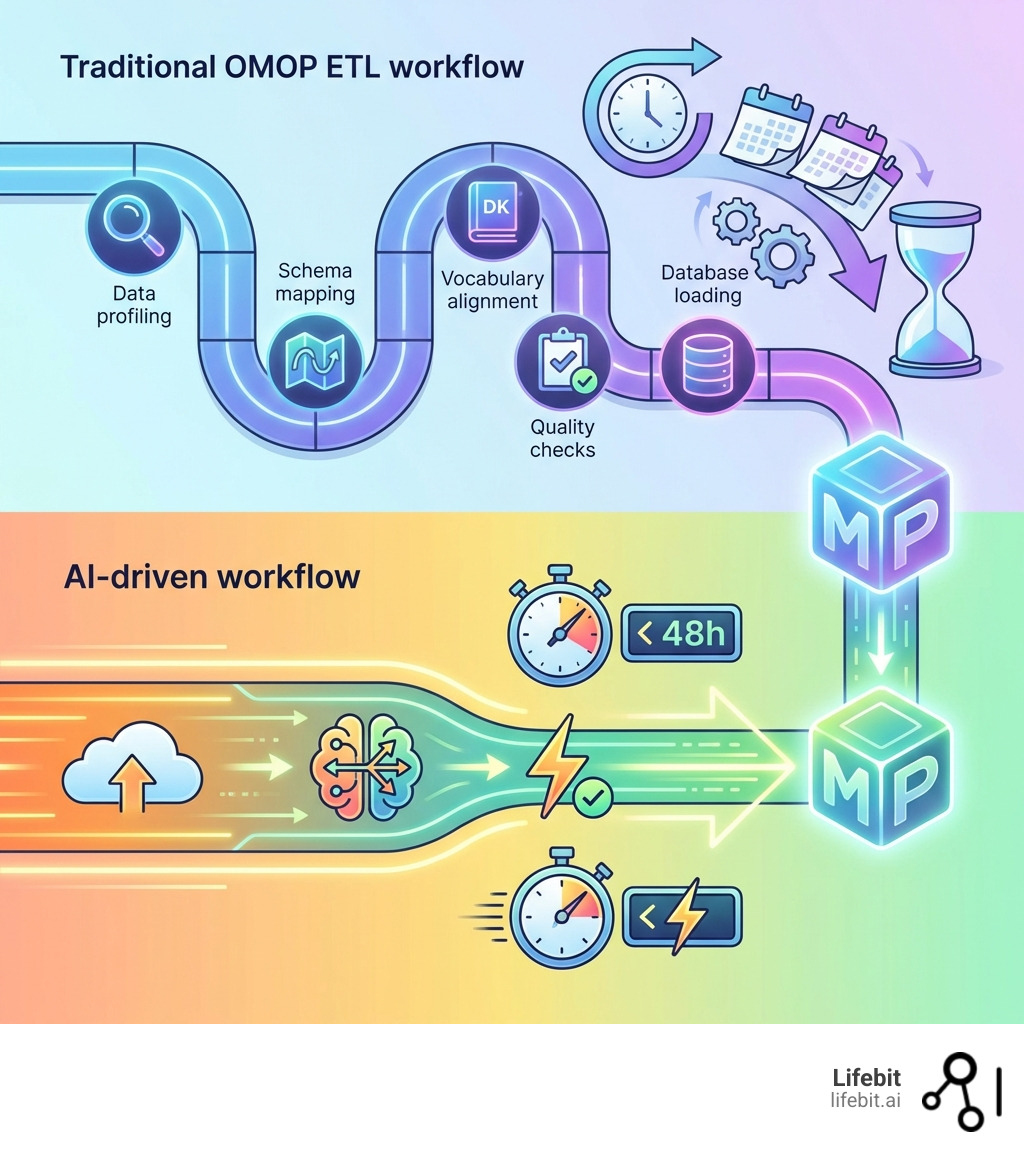
Slash Harmonization Time from Months to Minutes with AI for OMOP
Imagine converting complex, messy clinical data into a research-ready OMOP format in hours, not months. This is the reality AI for OMOP solutions can deliver. By leveraging Large Language Models (LLMs) and agentic AI workflows, platforms like Lifebit’s can automate data change, rapidly processing structured data from EHRs, FHIR, and claims, as well as the vast unstructured text and multimodal data that traditional methods miss.
Our federated AI platform automates data mapping, accelerating the conversion of source tables to OMOP. This yields research-ready data in under 48 hours, with mapping taking seconds, not weeks, in real-world deployments. For instance, we have supported projects mapping thousands of concepts at scale with automated quality metadata. This efficiency is transformative for organizations in Europe, the USA, Canada, Singapore, Israel, and beyond, who are eager to open up the value of their real-world data without delay.
Automating the ETL Process with AI
The Extract, Transform, Load (ETL) process is a notorious healthcare bottleneck. Generative AI revolutionizes it by automating complex data interactions, including generating SQL queries for epidemiological research. Instead of data engineers manually writing intricate scripts, AI can generate the necessary code to extract and transform data, dramatically streamlining the ETL pipeline.
Key AI Techniques for OMOP Transformation
Effective AI for OMOP transformation relies on a suite of sophisticated AI techniques working in concert to interpret and structure complex health data. These are not just isolated tools but interconnected components of an intelligent workflow that mirrors and accelerates expert human processes.
Named Entity Recognition (NER) for Clinical Data
The foundational step in understanding unstructured text is identifying what matters. Clinical NER models are specifically trained to scan clinical notes, pathology reports, and discharge summaries to pinpoint and categorize key pieces of information. For example, in the sentence, “Patient presents with persistent cough and was prescribed amoxicillin 500mg for suspected community-acquired pneumonia,” a clinical NER model would identify “persistent cough” as a SYMPTOM, “amoxicillin 500mg” as a TREATMENT (including dosage), and “community-acquired pneumonia” as a CONDITION. This foundational step turns a wall of text into a structured, machine-readable list of clinical concepts, ready for the next stage of analysis.
Relation Extraction to Build Clinical Context
Identifying entities is not enough; understanding their relationships is what creates meaning and ensures accuracy. Relation Extraction models analyze the output of NER to connect the dots. Using the previous example, it would establish relationships like: (amoxicillin 500mg) - treats - (community-acquired pneumonia). More importantly, it provides crucial context. It can distinguish between a patient’s current condition, family history (e.g., “father had heart disease”), a hypothetical concern (“rule out malignancy”), or a past condition (“history of asthma”). This contextual understanding prevents erroneous data mapping—like assigning a family member’s disease to the patient—and ensures the patient’s longitudinal story is captured accurately.
Automated Mapping to Standard Vocabularies (SNOMED, ICD, LOINC)
Once clinical concepts are extracted and contextualized, they must be translated into the standardized language of OMOP. This is where AI-powered mapping to vocabularies like SNOMED CT, ICD, LOINC, and RxNorm becomes critical. An AI model doesn’t just do a simple keyword search; it uses its understanding of clinical context to disambiguate terms. For instance, “cold” could refer to the common cold or a temperature sensation. The AI uses surrounding text to map it correctly to the SNOMED CT concept for “Common cold” (82272006). It also normalizes countless abbreviations, local jargon, and misspellings (e.g., mapping “T2DM” or “diabetis” to the standard concept for “Type 2 diabetes mellitus”). This process is automated and can be validated against resources like the Athena terminology browser, with a human-in-the-loop to review low-confidence mappings, ensuring both speed and accuracy.
Advanced De-identification of PII
Protecting patient privacy is non-negotiable. AI models are trained to identify and redact a wide range of Personally Identifiable Information (PII) from unstructured text, going far beyond just names and addresses. They can detect and remove dates of birth, phone numbers, medical record numbers, and even quasi-identifiers that could be used to re-identify a patient, such as a rare occupation or specific location. This is done intelligently to preserve clinical utility. For example, an exact date of service might be redacted, but the model can retain the time interval between two events (e.g., “three weeks later”), which is vital for research while upholding strict privacy standards like HIPAA and GDPR.
These techniques empower users to create cohorts and concept sets for analysis with unprecedented ease. Researchers can define complex patient populations using natural language, which AI translates into precise OMOP concept IDs and data filtering logic. AI also improves data quality reporting by proactively identifying inconsistencies during the transformation.
Open up the Complete Patient Story: Unifying All Data for Deeper Insights
The ultimate goal of data harmonization is to build a clinically complete patient data model. By integrating structured EHR, FHIR, and claims data with previously inaccessible unstructured notes and multimodal data, AI solutions create a holistic view of each patient’s journey. This end-to-end harmonization significantly improves accuracy for downstream use cases like risk adjustment, outcomes analysis, quality reporting, and decision support.
A unified model gives researchers and clinicians richer, more granular information for deeper insights and more reliable evidence. A platform designed for patient journeys can track a patient’s health trajectory across care settings, linking diagnoses, treatments, and outcomes from disparate sources. This ensures no critical piece of the patient’s story is missed.
Here is how AI-driven OMOP harmonization stacks up against traditional methods:
| Feature | Traditional OMOP Harmonization | AI-driven OMOP Harmonization |
|---|---|---|
| Speed | Months to years | Hours to days (research-ready data in under 48 hours, mapping in seconds) |
| Accuracy | Variable, prone to human error, information loss from unstructured data | High, AI reduces errors, consistent mapping, captures unstructured nuances |
| Cost | Very high (manual labor, specialized teams) | Significantly lower (automation, fewer manual hours) |
| Data Completeness | Limited (struggles with unstructured/multimodal data) | High (integrates structured, unstructured, and multimodal data) |
| Scalability | Low (manual processes do not scale well) | High (AI can process millions of observations efficiently) |
| Insights | Delayed, potentially incomplete | Near real-time, comprehensive, deeper |
Core Components of an AI-Powered Harmonization System
An effective AI-powered OMOP harmonization system is an integrated pipeline designed for complex, high-volume health data. Core components generally include:
- Data Ingestion Layer: Securely acquires data from diverse sources and formats, including structured EHR systems, FHIR endpoints, claims databases, and raw unstructured text or medical images.
- AI-powered De-identification and NLP Engine: Identifies and removes PII from all data types. The NLP engine then extracts clinical entities, attributes, and relationships from free-text notes.
- Automated Terminology Mapping Module: Automatically maps extracted concepts to standardized OMOP vocabularies (SNOMED, ICD, LOINC, RxNorm). LLMs and AI agents interpret medical context to suggest the most appropriate OMOP concept IDs, allowing for human-in-the-loop review.
- Data Quality and Validation Engine: AI continuously monitors data quality, detecting anomalies, inconsistencies, and missing data. This engine helps ensure the data is research-grade and compatible for loading into relational databases, augmenting tools like OHDSI’s Data Quality Dashboard.
- OMOP-compliant Relational Database Output: The harmonized, validated data is loaded into an OMOP CDM-compliant relational database, ready for analysis.
These components work together to provide a seamless solution that accelerates OMOP conversion and enriches the resulting dataset.
The Transformative Benefits of an AI-Driven OMOP Approach
Adopting an AI for OMOP strategy delivers far-reaching advantages that fundamentally change how organizations derive value from their healthcare data. The benefits extend beyond speed and cost savings to the very quality and depth of insights generated.
Unprecedented Accuracy and Clinical Completeness
The single greatest benefit is the creation of a more clinically complete and accurate patient data model. Traditional ETL processes that ignore unstructured data miss up to 80% of relevant clinical information, including lifestyle factors (e.g., smoking status, alcohol use), reasons for medication changes, or social determinants of health mentioned only in notes. By using NLP to extract and map this information, AI builds a high-fidelity patient profile. This leads to more reliable and reproducible research findings, as analyses are based on a comprehensive picture of the patient’s health rather than an incomplete, structured-only snapshot.
More Precise Risk Adjustment and Population Health Management
Risk adjustment models are essential for fair reimbursement and effective population health management, but their accuracy depends on the quality of the input data. Claims and basic EHR data often fail to capture the full burden of a patient’s illness. An AI-harmonized OMOP dataset includes granular details from clinical notes, such as the severity of a condition (e.g., “NYHA Class III heart failure”) or the presence of comorbidities not formally coded. Identifying patients with “uncontrolled diabetes” versus “stable diabetes” from physician notes allows for much more precise risk stratification, enabling health systems to allocate resources to the patients who need them most.
Deeper and More Robust Outcomes Analysis
Understanding treatment effectiveness requires linking interventions to outcomes with high precision. AI makes this possible at scale by extracting detailed clinical events—like the start and stop dates of a medication, dose adjustments, and patient-reported outcomes from notes. This allows researchers to conduct more robust analyses. For instance, a study could investigate not just whether a drug worked, but why it was discontinued (e.g., due to side effects vs. lack of efficacy), information that is rarely available in structured data but is critical for generating meaningful real-world evidence.
Automated and Reliable Quality Reporting
Healthcare organizations are required to report on a wide range of quality measures for regulatory compliance and care improvement. Manually abstracting this data from patient charts is a slow, expensive, and error-prone process. An AI-powered system can automate the identification and extraction of these quality metrics (e.g., HbA1c control levels, cancer screening dates) directly from the source data during the OMOP harmonization process. This ensures that reports are generated from a complete, up-to-date, and validated dataset, improving the accuracy of submissions to bodies like CMS and enabling near real-time internal monitoring of care quality.
Accelerated and Granular Cohort Creation
Identifying specific patient populations for clinical trials or observational studies is a major bottleneck in research. With traditional methods, this can take weeks of writing complex SQL queries and manually reviewing charts. AI-powered tools, often with natural language interfaces, revolutionize this process. A researcher can ask, “Find all female patients aged 50-65 diagnosed with HER2-positive breast cancer who have not received trastuzumab,” and the system can translate this into a precise query against the OMOP database in seconds. This capability is critical for advancing personalized medicine, accelerating drug discovery, and enabling rapid-cycle research on rare diseases.
Beyond the CDM: Are GenAI and Knowledge Graphs the Future of RWE?
The evolution of healthcare data models is continuous. The rise of Generative AI (GenAI) and Knowledge Graphs (KGs) raises a key question: could they make traditional CDMs like OMOP obsolete for analyzing distributed data networks? While instrumental, CDMs have complexities and limitations. GenAI and KGs offer a path toward more dynamic data structures and real-time insights, potentially changing the landscape of real-world evidence.
This shift is relevant for distributed data networks (DDNs), where data resides in many diverse formats. Current CDM implementations in DDNs are resource-intensive, requiring significant upfront ETL at each site. GenAI and KGs promise to analyze data directly in its original form, reducing costs, preserving data details, and enabling real-time insights for better patient outcomes and safer medication use.
How GenAI and KGs Could Make CDMs Obsolete
A perspective review in Ther Adv Drug Saf asks whether generative AI will make common data models obsolete in future analyses of distributed data networks perspective review on GenAI and CDMs. The argument centers on several key capabilities:
- Analyzing data in its original form: GenAI could interpret natural language queries and generate code to interact directly with raw data, bypassing the need for extensive, lossy changes into a CDM.
- Natural language querying: Researchers could ask complex questions in plain English, and a GenAI system would translate them into executable queries across diverse datasets, democratizing data access.
- Automated SQL query generation: GenAI can automate generating context-aware SQL queries for epidemiological research, increasing efficiency and facilitating code reuse across different DDNs.
- Enhancing ontological consistency: AI and KGs can dynamically harmonize mismatched terminologies. KGs can standardize semantics across heterogeneous data, while GenAI can help define and monitor phenotype consistency over time.
- Overcoming rigid crosswalks: GenAI can generate contextually appropriate mappings that go beyond the rigid structures of traditional crosswalks, allowing for more flexible and accurate data interpretation.
Combining GenAI’s power with KGs’ structured knowledge could create a “fourth generation” of distributed data network analysis. This new paradigm, with GenAI-enabled access, KGs for standardization, and automatic code generation, would reduce the operational burden on data providers and enable more agile, dynamic data analysis.
The Impact on Pharmacovigilance and Drug Safety
For drug safety and pharmacovigilance, the implications are significant. The dynamic analysis capabilities of GenAI and KGs offer several advantages:
- Real-time signal detection: Instead of waiting for lengthy ETL processes, GenAI can analyze incoming data streams in near real-time, enabling faster detection of adverse drug reaction (ADR) safety signals.
- Analyzing distributed data networks without a CDM: GenAI and KGs can facilitate the analysis of diverse RWD in its native format across DDNs, accelerating research while preserving data granularity.
- Reducing operational burden: By automating data harmonization and query generation, GenAI can significantly reduce the manual effort for pharmacovigilance studies, freeing up experts to focus on interpretation.
- Automating complex data interactions: AI can automate extracting specific adverse events from free-text reports and linking them with patient demographics and medications.
- Predicting unknown adverse drug reactions: KGs, combined with AI, show promise in predicting unknown ADRs by identifying potential relationships between drugs, diseases, and patient characteristics.
By embracing these technologies responsibly within secure federated environments, we can address current limitations, open up deeper insights from complex datasets, and ultimately support better patient outcomes and public health in the UK, USA, Europe, Canada, Singapore, Israel, and other regions where we operate.
Frequently Asked Questions about AI for OMOP
How does AI handle patient privacy and data security?
AI uses advanced de-identification techniques to remove Protected Health Information (PHI) before analysis. In a federated architecture, raw data never leaves its secure, local environment, helping ensure compliance with regulations like HIPAA and GDPR. Lifebit’s platform is built with enterprise-grade security, fine-grained access controls, and end-to-end encryption to maintain high standards of data protection.
Can AI completely replace human oversight in the OMOP ETL process?
No. AI dramatically accelerates and automates the process, but human-in-the-loop validation is crucial. Experts review AI-suggested mappings and data quality checks to ensure accuracy and clinical relevance. This collaboration lets AI handle repetitive tasks while experts focus on complex cases and critical decisions.
What is the main advantage of using AI over traditional OMOP tools?
The primary advantage is the ability to rapidly and accurately incorporate unstructured data, such as clinical notes that contain a large portion of relevant patient information, into the OMOP model. This creates a more complete and clinically rich dataset for research. Traditional tools struggle with unstructured data, leading to significant information loss. AI for OMOP closes this gap, providing a holistic patient view for deeper, more reliable real-world evidence.
The Future of RWE is AI-Powered: What’s Next?
The journey from siloed, messy healthcare data to actionable, near real-time insights has been long. For years, the promise of real-world evidence was constrained by the challenge of standardizing complex clinical data into formats like OMOP. The advent of advanced AI, including Large Language Models, agentic AI, and Knowledge Graphs, marks a pivotal turning point.
We are moving from an era of manual labor and delayed insights to one of automated data change and dynamic analysis. This shift enables researchers to analyze a more complete patient story, including previously inaccessible unstructured and multimodal data, at unprecedented speed. Achieving research-ready data in under 48 hours, unifying fragmented health data in days instead of months, is now achievable.
The future of observational research will increasingly rely on AI to improve ontological consistency, data interoperability, and automate the entire ETL process. We anticipate an evolution where data models become more adaptive and evidence generation more efficient. Federated learning will be critical for enabling secure, large-scale research across distributed data networks, allowing insights from vast datasets without compromising privacy.
At Lifebit, our federated AI platform is designed for this future. With components such as the Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence and Analytics Layer), we provide secure, real-time access to global biomedical and multi-omic data, with built-in capabilities for harmonization, advanced AI/ML analytics, and federated governance. This empowers biopharma, governments, and public health agencies in the UK, USA, Europe, Canada, Singapore, and Israel to open up the full potential of their RWD.
Ready to revolutionize your real-world evidence generation with AI for OMOP?

