Genomics England Explained: Pioneering the Future of Health

How Genomics England Ends the 5.6-Year Diagnostic Odyssey
Genomics England is a UK government-owned company established in 2013 to transform healthcare through genomic medicine. Here’s what you need to know:
- Mission: Integrate whole genome sequencing into routine NHS care to improve diagnosis and treatment
- Flagship Achievement: The 100,000 Genomes Project sequenced 100,000 genomes from NHS patients with rare diseases and cancer
- Current Scale: Supporting the NHS Genomic Medicine Service, which has sequenced over 100,000 additional genomes
- Data Infrastructure: Created the National Genomic Research Library, one of the world’s largest genomic databases with 50 petabytes of data
- Research Network: Enables 1,500+ researchers globally to access de-identified genomic data through secure environments
- Impact: Provided new diagnoses for thousands of rare disease patients who waited an average of 5.6 years for answers
For too many patients, getting a genetic diagnosis feels like wandering through a maze blindfolded. A child shows symptoms that baffle doctors. Tests come back inconclusive. Years pass. Families lose hope. This is the diagnostic odyssey—and it’s exactly what Genomics England was created to end.
By partnering with the NHS to sequence patients’ entire genomes and linking that data with clinical records, Genomics England is turning genomic science into practical healthcare. The results speak for themselves: actionable findings in around 50% of cancer participants and new diagnoses for thousands of families who had been searching for answers.
But this isn’t just about better diagnosis. It’s about building the data infrastructure that makes personalized medicine possible at population scale—securely, ethically, and in a way that benefits everyone.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, where we build federated data platforms that enable organizations like Genomics England to open up genomic insights while keeping data secure and compliant. Having worked in computational biology and precision medicine for over 15 years, I’ve seen how the right technology infrastructure can accelerate the journey from genome to treatment.
The 100,000 Genomes Project: How It Delivered Actionable Findings to 50% of Cancer Patients
Founded by the UK’s Department of Health and Social Care in July 2013 as a 65th birthday present for the NHS, Genomics England was created to bring genomic medicine from research into everyday healthcare. It represented a fundamental rethinking of how the NHS could diagnose and treat patients.
The mission was to partner with the NHS and integrate whole genome sequencing into routine clinical care. The goal was to make it a standard tool, not a luxury service, to help millions of patients get faster diagnoses, targeted treatments, and better outcomes.
As a government-owned company, Genomics England benefits from substantial funding through the Department of Health and Social Care, UK Research and Innovation, LifeArc, the National Institute for Health and Care Research, and the Wellcome Trust. This backing reflects a national commitment to making genomic medicine a reality for everyone, not just those who can afford it privately.
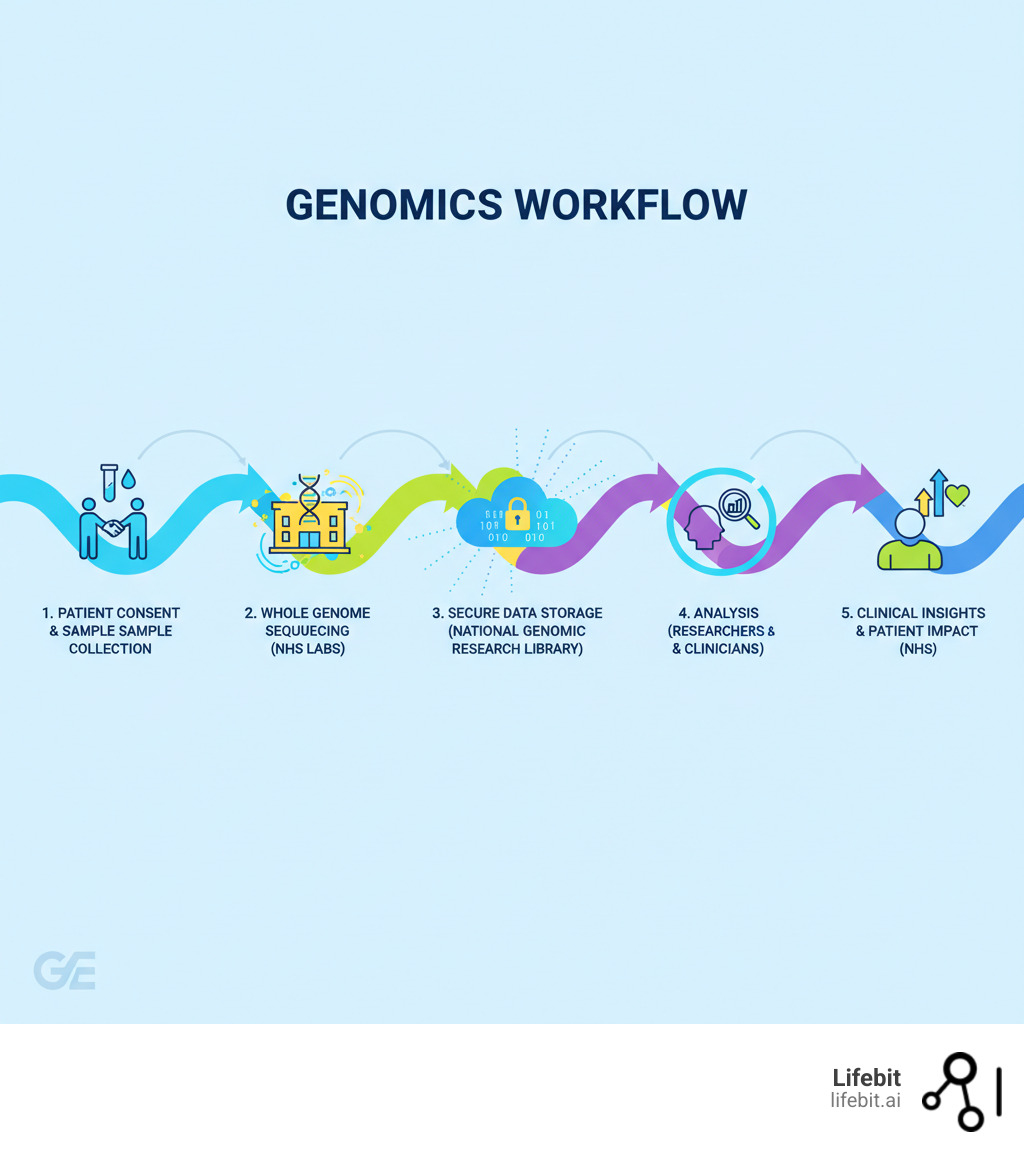
The partnership with the NHS goes far deeper than just running research studies. Genomics England provides the diagnostics technology and infrastructure that powers the NHS Genomic Medicine Service—a nationwide effort to deliver consistent, equitable genomic care to England’s 55 million people. This service, outlined in the NHS Long Term Plan and the Accelerating Genomic Medicine in the NHS Strategy, represents a fundamental shift in how the health service operates. You can learn more about the origins of Genomics England and how it evolved from an ambitious idea into a cornerstone of modern NHS care.
The Landmark 100,000 Genomes Project
The 100,000 Genomes Project put Genomics England on the map. Launched in 2013, this groundbreaking initiative set out to sequence 100,000 whole genomes from NHS patients living with rare diseases or cancer. The numbers alone tell an incredible story: 50 petabytes of data—roughly three times the entire Library of Congress—all containing the genetic blueprints of real patients seeking answers.
For families trapped in the diagnostic odyssey, this project was life-changing. After an average wait of 5.6 years for a diagnosis, thousands of participants finally got answers. Imagine spending years visiting specialists, undergoing test after test, watching your child struggle with unexplained symptoms—and then finally learning what’s actually wrong. That’s the power of whole genome sequencing when applied at scale.
The impact on cancer care was equally remarkable. Around 50% of cancer participants received actionable findings—insights that could directly inform their treatment decisions. This wasn’t theoretical research; these were real clinical insights that helped doctors choose the most effective therapies for individual patients.
But perhaps the project’s greatest achievement was proving that genomic medicine could work within a national healthcare system. It showed that whole genome sequencing wasn’t just scientifically valuable—it was clinically practical and ready for routine NHS care. The NHS Genomic Medicine Service, which has since sequenced over 100,000 additional genomes, exists because the 100,000 Genomes Project demonstrated what was possible. More on the 100,000 Genomes Project shows how this landmark study transformed from ambitious vision to healthcare reality.
Ethics and Governance: Building Patient Trust
When you’re working with people’s genetic information—arguably the most personal data that exists—trust isn’t optional. Genomics England built its entire operation around robust ethical principles and governance structures designed to protect participants while enabling groundbreaking research.
Every patient who participates provides informed consent after receiving clear, straightforward explanations about how their genomic and clinical data will be sequenced, stored, and used for research. No surprises, no fine print designed to confuse. Just honest conversations about what participation means.
Data security is treated with the seriousness it deserves. All genomic and clinical information is carefully de-identified before researchers can access it, stripping away personal identifiers while preserving the scientific value of the data. This allows researchers to make findies without ever knowing whose genome they’re studying.
What really sets Genomics England apart is the Participant Panel—a diverse group of people, many of whom participated in genomic studies themselves, who advise on everything from ethical questions to data access policies and overall strategy. These aren’t token representatives sitting in the corner of meetings. They’re active voices shaping how the organization operates, ensuring that patient perspectives inform every major decision.
An independent Ethics Advisory Committee provides additional oversight, tackling complex ethical dilemmas and ensuring that innovation never comes at the expense of participant welfare. Together, these governance structures create accountability at every level. The Participant Panel’s role demonstrates how patient involvement can be more than consultation—it can be genuine partnership in building the future of healthcare.
From 25 Hours to 23 Seconds: The Tech That Cut Genomic Analysis Time by 99%

The 100,000 Genomes Project generated a staggering 50 petabytes of data—an ocean of information roughly three times the entire Library of Congress. And that was just the beginning.
Each whole genome sequence contains billions of data points, creating big data challenges far beyond the capacity of traditional on-site servers. The sheer volume, speed, and analytical demands of modern genomics required a new approach.
Cloud computing provided the solution. Genomics England partnered with AWS to migrate its massive datasets and research environments to the cloud, fundamentally changing genomic research. Analysis tasks that once took researchers 25 hours now complete in just 23 seconds—a 99% reduction in processing time.
This isn’t just about speed for speed’s sake. When a child is suffering from an undiagnosed rare disease, every hour matters. When an oncologist is trying to identify the right treatment for a cancer patient, faster analysis can literally save lives. The high-performance computing infrastructure built on services like Amazon EC2 provides the elastic capacity needed to scale up for complex analyses and scale down during quieter periods.
And then there’s the question of durability. Genomic data represents an irreplaceable resource—you can’t just ask patients to provide samples again if something goes wrong. Amazon S3 storage offers 99.999999999% durability, which means this precious data is protected against loss with near-absolute certainty. For academic and government-funded research, the pay-as-you-go model also provides crucial flexibility, adapting to the realities of grant cycles and budget constraints.
The National Genomic Research Library
The National Genomic Research Library (NGRL) is where all this data comes together. Think of it as a centralized data resource that houses de-identified genomic data linked with clinical information from hundreds of thousands of NHS patients. It’s one of the world’s largest collections of its kind, and it’s designed with one purpose in mind: accelerating scientific findy.
But the NGRL isn’t just a massive hard drive somewhere. It’s an active, living research ecosystem. Over 1500 researchers from around the globe can access this treasure trove through a secure Research Environment, analyzing patterns, testing hypotheses, and making findies that would be impossible with smaller datasets.
This is where global collaboration becomes reality rather than aspiration. A researcher in Manchester can work with colleagues in Boston and Tokyo, all accessing the same rich dataset without ever compromising patient privacy. The de-identified data means individual identities are protected, while the clinical linkage provides the real-world context that makes research meaningful.
At Lifebit, we build the Trusted Research Environments that enable this secure collaboration. Our federated platform allows approved researchers to access sensitive data for approved projects, breaking down silos without compromising the security essential for handling genetic information.
For researchers interested in tapping into this resource, Genomics England provides detailed guidance on accessing the National Genomic Research Library.
The Technology Powering Research at Genomics England
Behind every breakthrough findy at Genomics England sits a sophisticated technological foundation. At its core are Trusted Research Environments (TREs)—secure, controlled spaces where researchers can analyze sensitive genomic and clinical data without ever moving it from its protected environment.
Think of a TRE as a secure room where you can work with highly confidential documents, but you can’t take copies out with you. Everything stays protected, but you can still read, analyze, and learn from what’s inside. This approach solves one of genomic research’s biggest challenges: how do you enable collaboration while maintaining absolute data security?
The scalable infrastructure supporting these environments includes advanced analytics tools, artificial intelligence (AI), and machine learning (ML) capabilities. These aren’t just buzzwords—they’re practical tools for processing vast amounts of genomic data and identifying patterns that human researchers might miss. When you’re looking at millions of genetic variants across thousands of patients, AI can spot subtle correlations that point toward new diagnoses or treatment approaches.
The transition to cloud-based solutions has been transformative. High-performance computing architecture now allows for complex analyses that would have been prohibitively slow or expensive just a few years ago. Researchers can run sophisticated models, test multiple hypotheses, and iterate quickly—all while working within secure, compliant environments.
Lifebit’s platform, with its Trusted Research Environment (TRE) and other components, provides the technological backbone for secure, real-time collaboration. This allows researchers to focus on discovery rather than data management.
This infrastructure facilitates scientific breakthroughs by making complex data analysis more efficient and accessible. It’s the difference between asking a question and waiting months for an answer versus getting insights in days or even hours. And in healthcare, that difference can change lives.
Screening Newborns for 200+ Treatable Diseases: Inside Genomics England’s Next Chapter
The 100,000 Genomes Project proved that genomic medicine could work at scale within the NHS. Now Genomics England is asking a bolder question: what if we could prevent illness before it starts?
Their future vision isn’t just about diagnosing disease faster—it’s about catching conditions early enough to change their course entirely. This shift from reactive to proactive healthcare is what drives their ambitious goal to sequence 5 million genomes within five years. With advanced techniques like long-read sequencing, they’re capturing genetic information that previous technologies simply couldn’t see.
These pioneering initiatives represent a fundamental change in how we think about healthcare. Instead of waiting for symptoms to appear, genomic insights could guide preventive care from the very first days of life. At Lifebit, we see this as the natural evolution of precision medicine—and we’re building the secure infrastructure that makes it possible at population scale.
The Generation Study: Screening Newborns for Rare Conditions
Imagine a routine newborn heel prick test revealing not just a handful of conditions, but over 200 rare, treatable genetic diseases. That is the promise of the Generation Study, Genomics England‘s groundbreaking research into newborn genome screening. With 25,000 babies enrolled, the study is already changing lives. For example, a baby named Freddie was diagnosed with hereditary retinoblastoma through screening, allowing for timely treatment that saved his sight.
Newborn screening could end the diagnostic odyssey for many families. Instead of years of uncertainty while irreversible damage occurs, early diagnosis enables interventions when they are most effective, giving children the healthiest possible start in life.
This approach also raises important ethical questions about psychological impacts and the balance between early detection and overtreatment. The Generation Study is carefully evaluating these considerations to ensure any future routine screening is implemented thoughtfully and ethically. You can learn more about this transformative program here: About the Newborn Genomes Programme.
How Genomics England is Tackling Cancer with Cancer 2.0
Cancer claims nearly 10 million lives worldwide every year. For many patients, treatment still feels like educated guesswork—try this drug, see if it works, move to the next one if it doesn’t. Precious time is lost, and not every patient has time to spare.
Genomics England‘s Cancer 2.0 initiative is working to change that equation entirely. By integrating advanced genomic technologies into cancer care, they’re building the foundation for truly personalized oncology—where treatment decisions are guided by the specific genetic makeup of each patient’s tumor.
At the heart of this initiative is long-read sequencing, a technology that provides a far more complete picture of cancer’s genetic complexity than traditional methods. Short-read sequencing can miss structural variations, complex rearrangements, and other genomic features that might be crucial for understanding why a particular cancer behaves the way it does. Long-read sequencing fills in these blind spots, revealing biomarkers that could predict treatment response or identify targetable vulnerabilities.
This deeper insight is especially critical for complex cancers that have historically been difficult to diagnose or treat. By applying multimodal data analysis—combining genomic information with clinical data, imaging, and other sources—Cancer 2.0 aims to move oncology beyond the one-size-fits-all approach. The goal is straightforward: match each patient to the treatment most likely to work for them, the first time. Learn more about this initiative here: The Cancer 2.0 Initiative.
The Diverse Data Initiative
Here’s an uncomfortable truth: most genomic research has focused overwhelmingly on people of European descent. This means the genetic variants we understand best, the diagnostic tools we’ve built, and the treatments we’ve developed all work most reliably for that population. Everyone else is left with a less accurate picture.
This isn’t just an ethical problem—it’s a scientific one. Genetic variation is richer and more complex across diverse populations, and by studying only a narrow slice of humanity, we’re missing crucial insights that could benefit everyone.
Genomics England is tackling this head-on through the Diverse Data Initiative, a focused effort to build a more inclusive dataset by actively recruiting participants from underrepresented ethnic backgrounds. The goal is to ensure that genomic healthcare works equally well for all ethnicities, not just some.
This commitment to addressing health inequality goes beyond fairness. A more diverse dataset leads to better science—more complete understanding of human genetic variation, more accurate diagnostic tools, and therapies that work effectively across populations. At Lifebit, we build federated platforms specifically designed to integrate diverse datasets from global partnerships while maintaining security and privacy. This kind of inclusive data infrastructure is essential for genomic medicine to truly fulfill its promise. Find out more about this critical work here: The Diverse Data Initiative.
Beyond these flagship programs, Genomics England is driving other crucial initiatives. The Rare Therapies Launchpad focuses on developing individualized therapies for children with rare conditions, offering hope where little existed before. And in partnership with the MHRA, they’re establishing a Yellow Card Biobank—a pioneering resource to understand how genetic makeup influences medication safety, advancing the entire field of pharmacogenomics.
These initiatives collectively represent the next frontier of genomic medicine: proactive, predictive, personalized, and equitable healthcare for everyone.
Conclusion
What started as an audacious birthday gift to the NHS has become something far more profound. Genomics England began with a simple but powerful idea: sequence 100,000 genomes and see what happens. The answer? Everything changed.
We’ve watched families finally get answers after years of uncertainty. We’ve seen cancer patients receive treatments custom to their unique genetic makeup. We’ve witnessed a research project transform into the beating heart of the NHS Genomic Medicine Service, where genomic sequencing is now part of routine care for millions of people.
The numbers tell part of the story—50 petabytes of data, 1500+ researchers collaborating globally, actionable findings in 50% of cancer participants. But the real story is in the diagnostic odysseys that ended, the treatments that worked when others failed, and the babies whose conditions were caught early enough to make all the difference.
Genomics England has proven that secure, federated data analysis isn’t just a technical achievement—it’s a lifeline. The National Genomic Research Library has become one of the world’s most valuable research resources precisely because it balances two competing needs: making data accessible enough to drive breakthroughs, while keeping it secure enough to maintain patient trust.
Looking ahead, the ambition only grows. Initiatives like the Generation Study for newborn screening, Cancer 2.0 for personalized oncology, and the Diverse Data Initiative for health equity are building the foundation for a healthcare system that treats each person as a unique individual, not a statistical average.
At Lifebit, our federated AI platform provides the secure, real-time infrastructure that powers initiatives like Genomics England. Our components, including the Trusted Research Environment (TRE), turn genomic data into clinical insights while upholding the highest standards of security and governance.
The work at Genomics England shows what’s possible with visionary leadership, patient trust, cutting-edge science, and the right technology. It’s not just about collecting data; it’s about connecting research to clinical care, enabling global collaboration, and ensuring the genomic revolution benefits every patient, regardless of their background.
The future of healthcare is being written right now, one genome at a time. And it’s more personalized, more proactive, and more promising than ever before.
Learn how federated technology powers large-scale genomic research.


