Medical Data Normalization: From Clinical Chaos to Terminology Triumph

Why Healthcare’s Data Crisis Demands Urgent Action
Medical data normalization is the systematic process of transforming fragmented, inconsistent clinical information from multiple sources—EHRs, labs, billing systems, pharmacies—into standardized, unified datasets that enable accurate analysis and decision-making across healthcare systems.
What Medical Data Normalization Does:
- Standardizes disparate data formats, terminologies, and coding systems into a common structure
- Eliminates duplicates, inconsistencies, and errors that threaten patient safety
- Enables interoperability between disconnected health systems and data silos
- Prepares raw clinical data for AI/ML algorithms, quality measures, and research analytics
- Applies recognized standards (SNOMED CT, LOINC, RxNorm, HL7 FHIR, OMOP) for semantic consistency
Why It Matters:
When data isn’t normalized, healthcare organizations face real patient safety risks. A single patient’s medications might appear as “1 tablet,” “one tab,” or “Tab 1x” across different systems. Lab results recorded in mmol/L at one facility won’t align with mg/dL at another. These inconsistencies lead to incorrect clinical decisions, failed quality measures, and research built on unreliable data.
The stakes are high: validation studies show that 99.9% of medication codes can map correctly when proper normalization is applied, but without it, critical information remains locked in isolated silos. Organizations struggle with slow data onboarding, poor data quality, and the inability to deploy AI at scale—all while regulatory bodies demand faster evidence generation and real-time pharmacovigilance.
The solution isn’t optional. Semantic interoperability through normalization is now the critical foundation for population health analytics, clinical decision support, and the secondary use of EHR data that powers modern healthcare research.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, with over 15 years of expertise in computational biology, AI, and health-tech. Throughout my career building federated genomics and biomedical data platforms, I’ve seen how Medical data normalization transforms global healthcare by enabling secure, compliant analysis across previously siloed datasets.
Related content about Medical data normalization:
What is Medical Data Normalization and Why Does It Matter?
At its core, Medical data normalization is about order. Imagine trying to run a global logistics company where half your team measures weight in pounds, the other half in kilograms, and some just write “heavy.” You’d have a disaster on your hands. In healthcare, this “disaster” happens every day in electronic health records (EHRs).
The concept of normalization isn’t new; it traces back to Edgar F. Codd, an IBM computer scientist who, in the 1970s, revolutionized how we think about databases. He proposed that data should be organized to reduce redundancy and improve data integrity. In the medical world, this means ensuring that a “Heart Attack” recorded in an ambulance report, a “Myocardial Infarction” in a hospital discharge summary, and an “ICD-10 I21.9” in a billing claim all point to the exact same clinical concept.
The Anatomy of Clinical Chaos
Without this process, we are left with “clinical chaos.” This chaos manifests in three primary ways:
- Syntactic Inconsistency: Different date formats (DD/MM/YYYY vs MM/DD/YY) or naming conventions for patients.
- Semantic Ambiguity: Using the same term for different things, or different terms for the same thing. For example, “Cold” could mean a common virus or a temperature sensitivity.
- Structural Fragmentation: Data stored in different tables or formats (JSON vs. SQL) that cannot be joined without significant manual effort.
Data normalization ensures that every piece of information—whether it’s a lab result, a medication dose, or a surgical procedure—is specific, expected, and transportable. It allows us to move beyond what is health data standardisation? and toward a future where data from a clinic in Singapore can be seamlessly compared with data from a research hospital in London.

The Critical Need for Semantic Interoperability
Interoperability is often discussed as a technical hurdle—can System A talk to System B? But the real challenge is semantic interoperability: do they actually understand each other?
The 21st Century Cures Act defines interoperability as the ability to exchange and use electronic health information without special effort. However, “Meaningful Use” (MU) requirements have shown us that simply having digital data isn’t enough. If the data is messy, it remains locked in “data islands.” Medical data normalization bridges these islands, translating varied terminologies into a common language. This is the “secret sauce” for high-quality results in any health data harmonization complete guide.
Benefits for Clinicians and Researchers
Why should a busy clinician care about database theory? Because normalized data saves lives.
- For Clinicians: It provides accurate patient representation. When a physician looks at a patient’s history, they see a unified timeline rather than a fragmented list of conflicting codes. This improves clinical decision support (CDS) and reduces the risk of medication errors. For instance, a normalized system can trigger an allergy alert even if the allergy was recorded in a different hospital using a different coding system.
- For Researchers: It enables population health analysis at scale. Instead of spending 80% of their time cleaning “messy” data, researchers can jump straight to analysis. This is particularly vital for rare disease research, where data must be aggregated from dozens of global sites to reach statistical significance.
- For Administrators: It streamlines compliance and reporting. Automated quality measures (like those from the National Quality Forum) become possible only when data is normalized. This reduces the administrative burden of manual chart reviews.
We’ve seen that these seven benefits of health data standardisation lead to lower costs and better patient outcomes by ensuring that the right data reaches the right person at the right time.
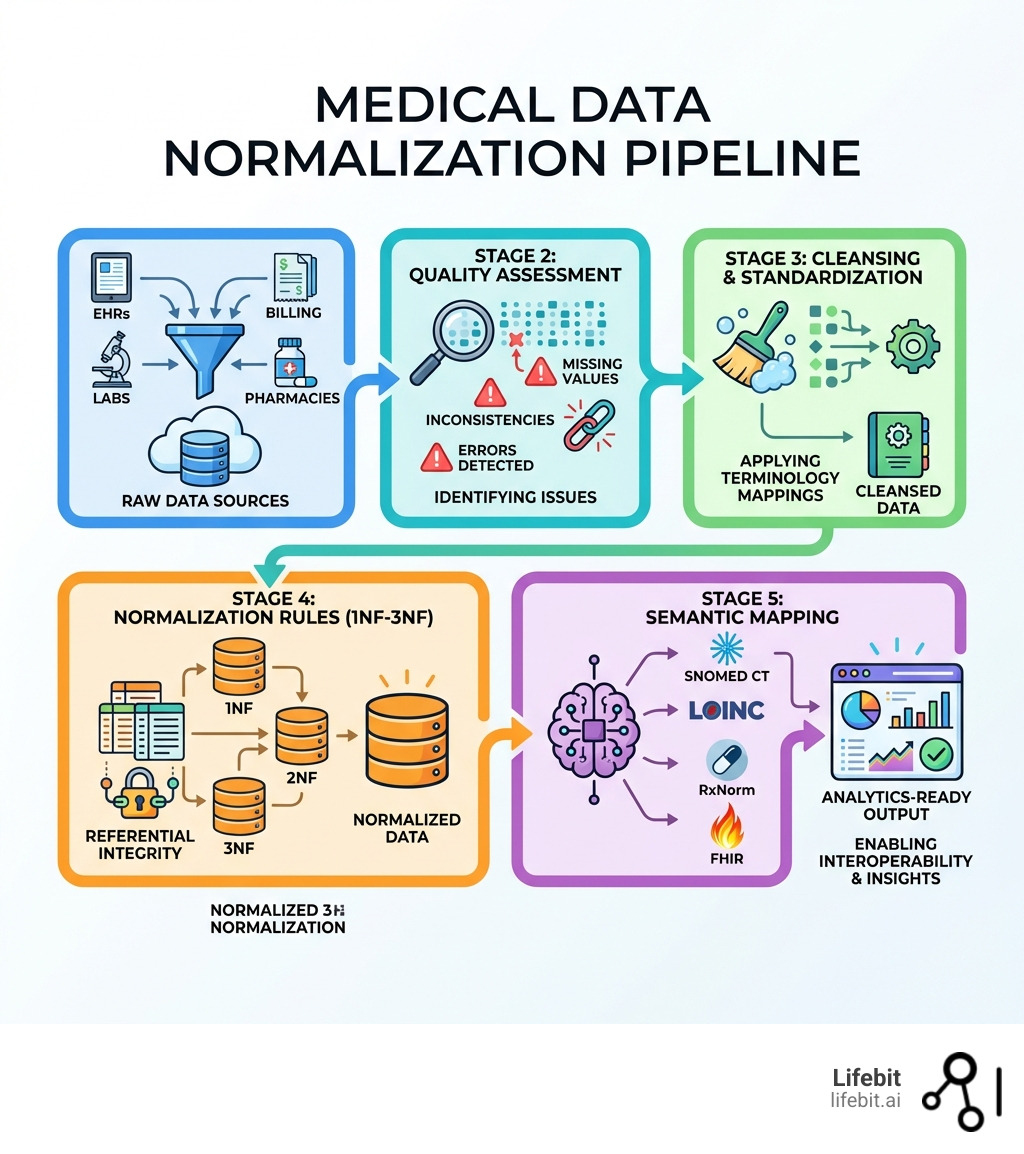
The 5-Step Process: From Raw Input to Standardized Output
Normalizing medical data isn’t a “one-and-done” task; it’s a systematic pipeline. To get from a messy EHR export to a research-ready dataset, we follow a rigorous workflow that combines database theory with clinical expertise.
Step 1: Data Extraction and Cleansing
The first step is gathering data from disparate sources—EHRs, labs, and even wearable devices. This raw data is usually “messy,” containing noise, outliers, and duplicates. We use tools like the MITRE Identification Scrubber Tool (MIST) to de-identify data for HIPAA compliance and “scrub” the records.
This involves:
- Reformatting dates: Converting all entries to ISO 8601 (YYYY-MM-DD).
- Unit Conversion: Ensuring all weights are in kilograms and all heights in centimeters to prevent calculation errors in BMI or drug dosing.
- Deduplication: Identifying if “John Doe” and “J. Doe” at the same address are the same patient.
This health data standardisation end-to-end analysis ensures that the foundation of our database is clean.
Step 2: Applying Normalization Rules (1NF to 3NF)
This is where we apply Edgar F. Codd’s database rules to clinical tables:
- First Normal Form (1NF): Eliminate duplicate columns and ensure each cell contains only one value. No more “Diagnosis: Asthma; Diabetes” in a single box. Each diagnosis gets its own row.
- Second Normal Form (2NF): Ensure all data in a table relates directly to the primary key. For example, in a ‘Visits’ table, the ‘Clinic Address’ should not be repeated for every visit; it should live in a separate ‘Clinics’ table linked by a Clinic ID.
- Third Normal Form (3NF): Remove columns that don’t rely solely on the primary key. This reduces redundancy to the absolute minimum, ensuring that if a patient’s phone number changes, it only needs to be updated in one place.
These rules ensure referential integrity, meaning your data remains consistent even as you update it. It’s a vital part of data harmonization techniques used to prevent anomalies.
Step 3: Terminology Mapping and Transformation
The final “triumph” is mapping. We take proprietary local codes (like “L-102” for a glucose test) and map them to international standards like LOINC. This is where data harmonization services become essential, as they handle the labor-intensive task of matching millions of local entries to their standardized equivalents.
We must also navigate data harmonization overcoming challenges, such as:
- Post-coordination: When a single clinical concept requires multiple codes (e.g., “Fracture of the left femur”).
- Mapping Ambiguity: When a local code is too vague to map directly to a specific SNOMED CT term.
- Version Control: Ensuring that mappings are updated as new versions of ICD or LOINC are released.
Essential Standards and Technologies for Normalizing Medical Data
To achieve true Medical data normalization, we rely on a “alphabet soup” of standards. Each serves a specific purpose in the clinical ecosystem:
- SNOMED CT: The most comprehensive clinical terminology for diagnoses, symptoms, and findings. It uses a poly-hierarchical structure, allowing a “Lung Infection” to be classified under both “Respiratory Disease” and “Infectious Disease.”
- LOINC: The gold standard for identifying laboratory observations and measurements. It defines tests based on six axes: Component, Property, Time, System, Scale, and Method.
- RxNorm: The essential system for normalized names for clinical drugs. It links NDCs (National Drug Codes) to a unified concept, allowing systems to recognize that two different brands of the same medication are identical.
- ICD-10-CM/PCS: Used primarily for billing and epidemiological tracking. While less granular than SNOMED, it is the global standard for mortality and morbidity reporting.
Research projects like the SHARPn consortium have demonstrated that using these standards in a unified informatics infrastructure is the only way to achieve high-throughput phenotyping.
Natural Language Processing (NLP) and Unstructured Data
Roughly 80% of medical data is unstructured—think doctor’s notes, pathology reports, and discharge summaries. To normalize this, we use AI for data harmonization.
Technologies like Apache cTAKES (built on the UIMA framework) allow us to perform named entity recognition. We can identify a mention of “shortness of breath” in a note and automatically map it to the correct SNOMED CT code, even accounting for negation (e.g., “patient denies shortness of breath”). Advanced NLP models can now also extract temporal relationships, identifying whether a symptom occurred before or after a specific medication was started.
Information Models and Frameworks
Beyond simple codes, we need models that describe how data elements relate. Clinical Element Models (CEMs) and the Quality Data Model (QDM) provide the grammar for clinical logic. For instance, the QDM helps define exactly what constitutes a “diabetic patient” for a quality report—specifying the required diagnosis codes, lab values (A1c > 6.5%), and medication history.
Modern systems are increasingly moving toward HL7 FHIR (Fast Healthcare Interoperability Resources). FHIR uses “Resources” (like Patient, Observation, or Medication) and modern RESTful APIs to allow for structured data exchange. Following a data integration standards healthcare guide is critical for ensuring these models work together across different software vendors.
Why Medical Data Normalization is the Foundation for AI and ML
You’ve heard the phrase “garbage in, garbage out.” In AI, this is a literal truth. If you train a machine learning model on unnormalized data, it will learn the inconsistencies of the data rather than the patterns of the disease. For example, if one hospital records “Type 2 Diabetes” and another uses “T2DM,” an unnormalized model might treat these as two different conditions, leading to skewed results and “algorithmic bias.”
Medical data normalization provides the “clean” training data required for:
- Predictive Modeling: Forecasting which patients are at risk of hospital readmission or sepsis before symptoms become severe.
- Early Detection: Identifying subtle trends in rising glucose levels or declining kidney function across thousands of patients to prevent the onset of chronic disease.
- Precision Medicine: Matching complex genetic profiles to the most effective therapies by ensuring that phenotypic data (symptoms and outcomes) is as structured as the genomic data.
This is why we view normalization as a clinical data harmonisation: trusted data factory for AI readiness.
The Role of Medical Data Normalization in High-Throughput Phenotyping
Phenotyping is the process of identifying a cohort of patients who share a specific condition or characteristic. In the past, this required manual chart review. Using normalized data, we can run “high-throughput” queries that identify thousands of patients in seconds.
By using rules engines like JBoss Drools, we can translate complex clinical criteria into executable scripts. For example, a phenotype for “Treatment-Resistant Hypertension” might require:
- Three different antihypertensive medications in the pharmacy record.
- A systolic blood pressure reading >140 mmHg recorded after 6 months of treatment.
- The absence of secondary causes like renal artery stenosis.
This allows for harmonizing disparate electronic health records across entire nations for large-scale clinical trials.
Real-World Success: Medical Data Normalization in Clinical Quality Measures
The impact of this work isn’t theoretical. In a study involving the Mayo Clinic, researchers used a normalization pipeline to execute the NQF 0064 quality measure (Diabetes: LDL Management).
- The Result: On a cohort of 273 patients, the platform successfully identified the exact numerator and denominator needed for the measure.
- The Success Rate: 1,000 HL7 medication messages were validated, with a 99.9% success rate in mapping to RxNorm.
- The Impact: This proved that automated, standardized quality reporting is not only possible but highly accurate. These are the kinds of results we aim for when from chaos to clarity: implementing OMOP for DHA data.
Normalization vs. Denormalization: Strategic Data Management
While normalization is the “gold standard” for data integrity and transactional systems, it’s not always the fastest way to read data for massive analytics. In “Big Data” and data warehousing, we sometimes use denormalization strategically.
| Feature | Normalization (1NF-3NF) | Denormalization |
|---|---|---|
| Primary Goal | Reduce redundancy & ensure integrity | Increase query & read speed |
| Data Structure | Many small, related tables (Snowflake Schema) | Fewer, large tables with redundant data (Star Schema) |
| Best For | Transactional systems (EHR data entry, OLTP) | Analytics & Data Warehousing (OLAP) |
| Integrity | High (updates happen in one place) | Lower (must update multiple locations) |
| Storage Space | Optimized/Minimal | Increased due to redundancy |
When to Choose Normalization
We choose normalization for transactional systems where data is frequently updated. It prevents “update anomalies”—for example, if a patient changes their address, you only want to update it in one table, not fifty. If the data were denormalized, you might update the address in the ‘Visits’ table but forget to update it in the ‘Billing’ table, leading to inconsistent records. This helps navigate health data standardisation technical challenges by keeping the “source of truth” clear.
When to Choose Denormalization
For high-speed analytics and reporting, we might “flatten” the data. If a researcher needs to query 100 million records to find a specific drug-drug interaction, a normalized database would require complex “joins” between dozens of tables, which can be computationally expensive and slow.
A denormalized “Data Lakehouse” or OLAP cube will return results much faster by pre-joining these tables. Understanding beyond integration: understanding data harmonization means knowing when to prioritize integrity and when to prioritize speed.
Many modern researchers use the OMOP Common Data Model (CDM), which strikes a balance. It uses a standardized, normalized schema for the underlying data but allows for the creation of “derived tables” that are optimized for specific analytical tasks. This approach ensures that the data remains scientifically rigorous while still being accessible for rapid discovery. You can learn more in our OMOP complete guide.
Frequently Asked Questions about Medical Data Normalization
What is the difference between syntactic and semantic normalization?
Syntactic normalization deals with the structure and format of the data. It ensures dates are in the same format (YYYY-MM-DD), that files use the same XML/JSON schema, and that character encoding (like UTF-8) is consistent. Semantic normalization deals with the meaning of the data. It ensures that “Type 2 Diabetes,” “T2DM,” and “Non-insulin dependent diabetes” are all mapped to the same SNOMED CT code (44054006). You need both for true interoperability.
How does normalization improve patient safety?
It eliminates ambiguity in critical care settings. If a pharmacist sees “one tab” and the system interprets it as “10 tablets” due to a coding error or lack of unit normalization, the result could be fatal. Medical data normalization ensures that dosages, units of measure, and allergy alerts are consistent across every care setting. It also ensures that a patient’s “Problem List” is accurate, preventing doctors from prescribing medications that might conflict with an existing condition recorded in a different system.
Why is RxNorm essential for medication data?
RxNorm provides a common language for clinical drugs and drug packs. Because drug manufacturers use different names, strengths, and National Drug Codes (NDCs) for the same generic medication, RxNorm acts as a “universal translator.” It allows systems to check for drug-drug interactions and perform medication reconciliation even if the medications were prescribed by different doctors using different EHRs. It is the backbone of modern e-prescribing.
Does normalization require manual data entry?
While manual mapping was common in the past, modern normalization relies heavily on automated ETL (Extract, Transform, Load) pipelines and AI-driven mapping tools. However, a “human-in-the-loop” is often still required for quality assurance, especially when dealing with highly complex or rare clinical concepts that automated tools might misinterpret.
What is the cost of not normalizing medical data?
The costs are both financial and clinical. Financially, organizations waste millions on manual data cleaning and redundant tests caused by missing information. Clinically, the cost is measured in medical errors, delayed diagnoses, and the inability to participate in cutting-edge clinical trials that require structured data for patient matching.
Cut Your Data Onboarding Time by 80% with Lifebit
The journey from clinical chaos to terminology triumph is complex, but you don’t have to walk it alone. At Lifebit, we’ve built the next generation of federated AI infrastructure to handle the heavy lifting of Medical data normalization for you.
Our platform—featuring the Trusted Data Lakehouse (TDL) and our R.E.A.L. (Real-time Evidence & Analytics Layer)—is designed to harmonize multi-modal, multi-omic data where it lives. By using federated governance, we enable secure, real-time access to global data without the need for risky data transfers.
Whether you are a public health agency in Singapore or a biopharma giant in New York, our Lifebit data standardisation tools ensure your data is research-ready, compliant with ISO 27001, GDPR, and HIPAA, and primed for the next wave of AI-driven discovery.
Stop struggling with messy data. Start accelerating your research.

