Cracking the Code: Your Guide to the OMOP Common Data Model

Drowning in EHR Chaos? Turn It into FDA-Ready Evidence with OMOP—No Data Movement
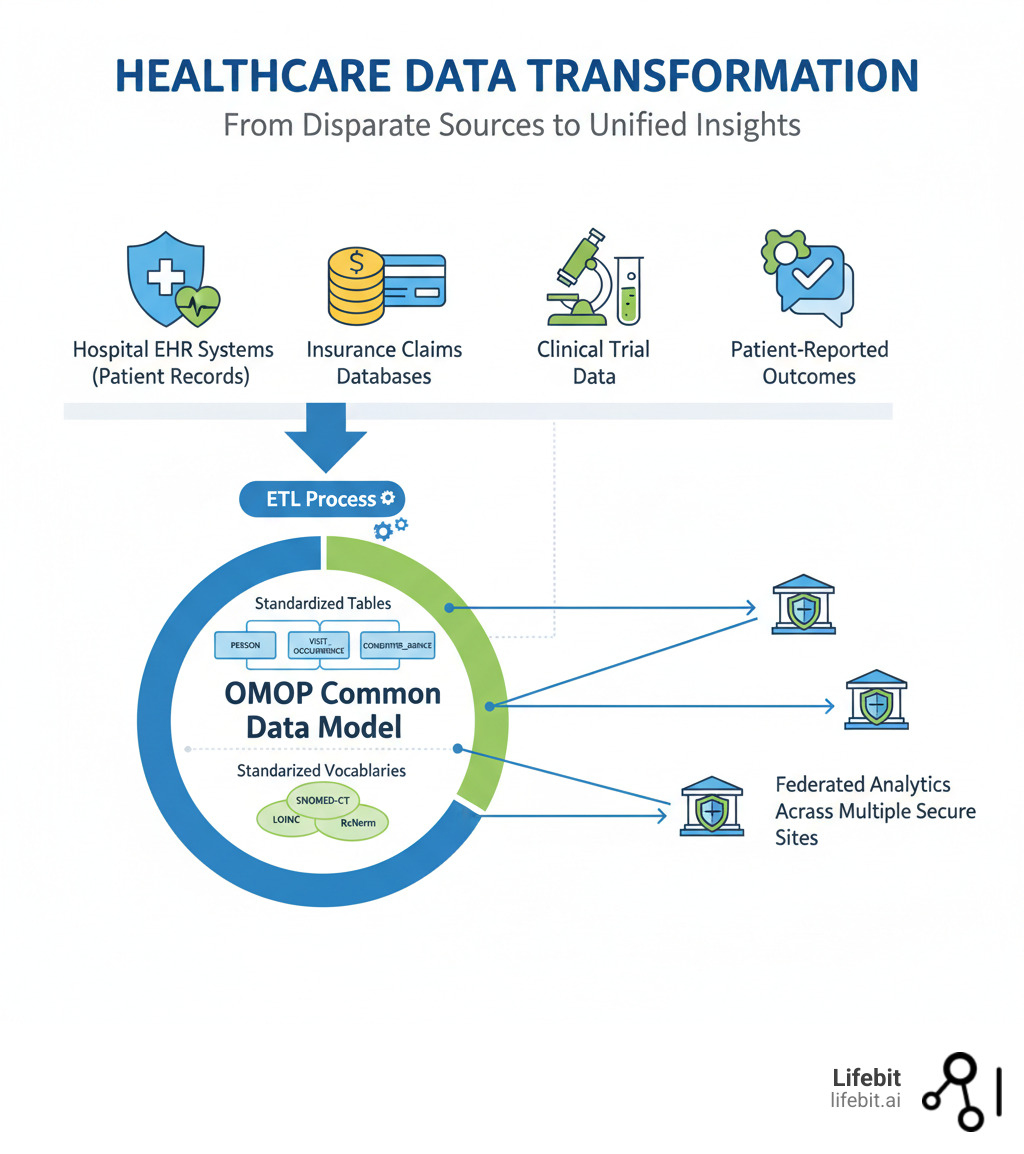
OMOP (Observational Medical Outcomes Partnership) is an open community data standard that transforms disparate healthcare data into a common format. This enables researchers to systematically analyze observational databases from hospitals, insurers, and health systems worldwide. The OMOP Common Data Model (CDM) standardizes both the structure and content of health data, allowing the same analytical tools to work across different datasets without moving sensitive information.
OMOP standardizes data structure into a consistent format and harmonizes terminologies by mapping various coding systems (like ICD and SNOMED) to a standard vocabulary. This enables federated analysis, allowing researchers to run the same query across multiple institutions while data remains secure at its source. The result is accelerated research, changing lengthy data integration projects into reusable analyses and generating Real-World Evidence (RWE) accepted by agencies like the FDA and EMA.
Healthcare organizations worldwide face a fundamental problem: their data is trapped in silos, using different formats and coding systems. This chaos makes large-scale research slow, expensive, and unreliable. The OMOP CDM solves this by acting as a universal translator for health data. Instead of forcing organizations to share raw data, OMOP transforms local data into a common language once, then enables standardized analytics to run anywhere. This approach powers collaborative research networks like the All of Us Research Program and major oncology initiatives.
I’m Maria Chatzou Dunford, CEO of Lifebit. We build federated platforms for secure analysis of OMOP-standardized data, and we’ve seen how it open ups the potential of biomedical data at scale.
Simple guide to omop terms:
Stop Burning Months on Data Wrangling: OMOP CDM Lets You Run the Same Study Across Hospitals This Quarter
Healthcare data from electronic health records, insurance claims, and patient registries holds immense potential, but it’s often unusable at scale. Different hospitals use different coding systems, labs use different units, and medications appear under different names. This data heterogeneity problem creates chaos that prevents us from open uping the true potential of healthcare data.
The OMOP Common Data Model is a philosophy for approaching observational health data. It creates a universal language that every healthcare system can speak, enabling systematic analysis and reproducible research across previously incompatible datasets. When data shares a common format and common representation, we can build a true Learning Healthcare System. This is a cyclical process where routine clinical data is used to generate evidence, and that evidence is fed back to clinicians to inform their practice, leading to improved patient outcomes. This continuous loop of data-to-evidence-to-practice is only possible when data is standardized and ready for analysis. This enables large-scale analytics that can answer questions no single institution could tackle alone. More info about OMOP is available on our blog.
The Problem and Solution
Imagine studying a new diabetes medication across multiple hospitals. Each source stores information differently. Electronic Medical Records are optimized for clinical care, while administrative claims data are built for provider reimbursement. This results in fundamentally different data structures and terminologies. For example:
- Semantic Heterogeneity: A blood glucose measurement might be recorded with different codes (LOINC, SNOMED CT, local proprietary codes) and units (mg/dL, mmol/L) at each site. A diagnosis of “Type 2 Diabetes” could be recorded using ICD-9, ICD-10, or a free-text note.
- Structural Heterogeneity: Hospital A might record every patient interaction as a single, long “encounter,” while Hospital B creates separate “visits” for the ER, inpatient stay, and follow-up consultation. One system might store lab results in one table and vital signs in another, while a second system combines them.
This forces researchers to spend months on custom data integration for every new study, writing bespoke code for each data source. This makes research slow, expensive, and limited in scope. The OMOP CDM solves this by providing a standardized structure and standardized content. The structure is a patient-centric relational database model that organizes clinical information consistently, regardless of its source.
The real power, however, comes from OMOP‘s comprehensive vocabularies, which ensure semantic consistency. These vocabularies map all source codes ICD-10, SNOMED CT, LOINC, RxNorm to standard concepts. This means “diabetes” is always represented the same way, no matter where the data originated.
This standardization enables federated analysis, a core capability of the OMOP ecosystem. Instead of centralizing sensitive patient data, analytical queries run at each data source, and only aggregate results are shared. This allows you to analyze data where it lives while preserving privacy. At Lifebit, our platform is built on this federated approach, allowing organizations to run analyses across global networks in days instead of months.
Avoid ETL Rework: The OMOP Tables and Mappings You Must Get Right First
Think of the OMOP CDM as a blueprint for a patient’s healthcare journey. It’s a relational database designed with logical consistency, so whether you’re tracking a single patient’s blood pressure or analyzing prescription patterns for millions, the data is organized in a predictable way. This makes complex analyses surprisingly straightforward.
To get there, real-world data goes through an ETL (Extract, Transform, Load) process. Data is extracted from source systems (like an EHR or claims database), transformed by mapping it to OMOP‘s structure and vocabularies, and then loaded into the OMOP database. This mapping process requires both technical and clinical expertise to ensure data integrity. At Lifebit, we’ve built tools to simplify this challenge, helping organizations map their data into OMOP efficiently.
Key Components and Structure
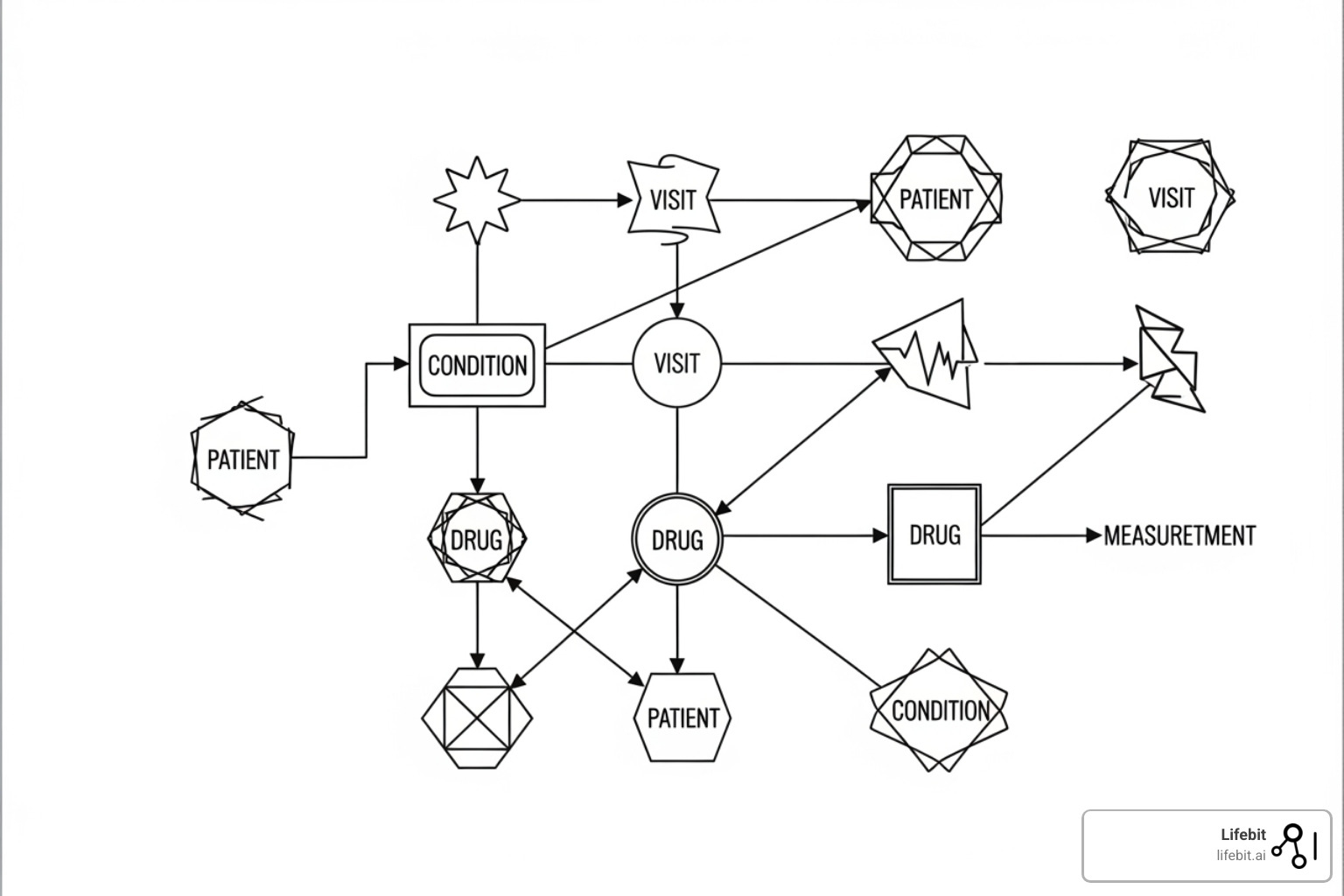
The OMOP CDM organizes information into interconnected tables. At its core, the PERSON table stores patient demographics. All other data connects back to this central identity.
- Clinical event tables capture what happens to patients. These include VISITOCCURRENCE (healthcare encounters), CONDITIONOCCURRENCE (diagnoses), DRUGEXPOSURE (medications), PROCEDUREOCCURRENCE (treatments), and MEASUREMENT (lab results).
- Health system tables provide context, such as LOCATION, CARE_SITE (facilities), and PROVIDER (clinicians).
- Derived elements like DRUGERA and CONDITIONERA group consecutive events, simplifying analyses of long-term treatment patterns or chronic diseases.
Let’s walk through an example: A 65-year-old male goes to the emergency room with chest pain.
- A new record is created in the PERSON table for this patient.
- The ER visit generates a record in VISIT_OCCURRENCE, linked to the patient’s
person_id. - He is diagnosed with an acute myocardial infarction (heart attack). This creates a record in CONDITION_OCCURRENCE, linked to the visit and the person.
- An ECG is performed. This is recorded in PROCEDURE_OCCURRENCE.
- His troponin levels are tested. The result (“0.8 ng/mL”) is stored in the MEASUREMENT table.
- He is given aspirin. This is captured in DRUG_EXPOSURE.
Each of these records is linked together, creating a detailed, longitudinal view of the patient’s journey that can be easily queried. This structure is based on decades of research into how observational data is used. For complete technical specifications, you can read more about the OMOP Common Data Model in the official documentation.
The Importance of Standardized Vocabularies
Standardized vocabularies are what make OMOP truly powerful. A common structure is meaningless if “heart attack” is coded five different ways. The vocabularies solve this by mapping every clinical concept diagnosis, medication, procedure, or measurement to a standard concept ID.
Your source data might use ICD-10-CM code I21.3 for “ST elevation (STEMI) myocardial infarction,” while another uses SNOMED CT code 233837004. During the ETL process, both of these source concepts are mapped to a single standard concept in the OMOP vocabulary (e.g., SNOMED concept ID 410429000 for “Acute myocardial infarction”). This translation layer is the key to semantic interoperability. When you query for “acute myocardial infarction” using the standard concept ID, you automatically capture every instance, regardless of the original code. This semantic consistency allows you to define a cohort like “patients with Type 2 diabetes taking metformin” and run that exact definition across datasets in Boston, Berlin, and Beijing, knowing the results will be comparable.
For researchers, the Athena vocabulary browser is an essential tool for exploring these vocabularies and finding the correct standard concepts for their analyses. To dive deeper, you can read more about OHDSI’s standardized vocabularies.
Proof It Works: OMOP Delivers Global RWE Regulators Trust—With Zero Data Movement
The OMOP CDM has moved from theory to practice, becoming the backbone for some of the world’s most ambitious healthcare research initiatives. Its power lies in enabling collaborative research at scale. When hundreds of institutions speak the same data language, network studies spanning continents become feasible. A researcher can design a study once, and colleagues across the globe can run the exact same analysis on their local dataall without a single patient record crossing borders.
This global evidence generation is changing how we understand disease and treatment effectiveness. Real-World Evidence (RWE) from OMOP-standardized data is now routinely accepted by regulatory agencies like the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for post-market surveillance and other decisions.

By enabling rapid, large-scale analysis, OMOP helps researchers identify unmet patient needs faster, optimize treatment pathways, and develop personalized medicine. Questions that once took years to answer can now be tackled in months or weeks.
Case Studies in Action
Global initiatives demonstrate OMOP’s impact. The European Health Data and Evidence Network (EHDEN) is a flagship project aiming to standardize the health records of over 100 million EU citizens to the OMOP CDM. By creating a federated network of data partners, EHDEN has enabled rapid, large-scale observational studies, including critical research on COVID-19 treatment effectiveness and vaccine safety during the pandemic. In oncology, projects like ONCOVALUE, EUCAIM, and CHAIMELEON are creating massive, analyzable evidence repositories from previously unstructured data, helping to accelerate the development of personalized cancer therapies. Learn more at Oncovalue.org about how this work informs clinical care.
In the US, the All of Us Research Program, with over one million participants, and the Veterans Health Administration both built their data infrastructures on the OMOP CDM to ensure consistency and enable large-scale research. You can learn more about how the standard is used in the What is OMOP? All of Us Research Hub FAQ.
OMOP in Pharmacovigilance
A powerful application of the OMOP network is in pharmacovigilance—the science of monitoring the safety of medicines. Traditionally, detecting rare side effects relies on spontaneous reporting systems, which can be slow and incomplete. With a federated OMOP network, researchers can proactively search for safety signals. For example, if a new drug is suspected of causing a rare liver injury, a query can be run across the network to compare the incidence of liver injury in patients taking the new drug versus a similar, established drug. Because the analysis runs on millions of patient records, even rare events can be detected with statistical confidence, providing regulators with crucial safety information much faster than traditional methods.
The Benefits of Adopting the OMOP CDM
The global momentum behind OMOP reflects tangible benefits for all stakeholders.
- For Researchers: Analysis is faster, tools are reusable, and statistical power increases dramatically. Instead of spending 80% of their time on data preparation, they can focus on scientific findy, not data wrangling.
- For Organizations: Data transforms from a compliance burden into a strategic asset. Hospitals improve data quality and can benchmark their outcomes against peers. Pharmaceutical companies can participate in large-scale network studies, accelerating drug development and fulfilling post-market safety commitments.
- For Patients: Faster research means more effective treatments and better outcomes. Large-scale analysis enables truly personalized medicine, creating a healthcare system that continuously learns and improves from every patient’s experience.
Get Productive Fast: OHDSI’s Free Tools That Cut Weeks Off Your OMOP Build
The OMOP Common Data Model is the heart of a global community called Observational Health Data Sciences and Informatics (OHDSI). OHDSI is an open-source, interdisciplinary collaborative that maintains and evolves the OMOP CDM, fosters its adoption, and develops a suite of free tools to leverage standardized data.
The OMOP CDM is a living model, currently on version v5.4, with refinements driven by community feedback. The OHDSI CDM Working Group oversees this evolution, ensuring the model remains robust and responsive to real-world research needs.
The OHDSI Toolbox: Open-Source Power
OHDSI provides a powerful ecosystem of open-source tools. ATLAS is a web-based application for designing and executing observational analyses without writing code. Other key tools include the Data Quality Dashboard, which runs over 3,500 automated checks to ensure data integrity, and White Rabbit, which assists in the initial ETL mapping process. These tools are freely available, well-documented, and constantly improved by the community.
How to Get Started
Adopting OMOP is a manageable journey with the right approach. The process can be broken down into four key phases:
- Assess Your Data (Scoping & Profiling): Before writing any code, thoroughly analyze your source systems. Use tools like OHDSI’s White Rabbit to scan your database schemas and profile the content. This phase answers critical questions: What data do you have? Where does it live? What coding schemes are used (ICD, LOINC, local codes)? How complete is the data? This initial assessment is the foundation for your project plan and resource allocation.
- The ETL Process (Mapping & Transformation): This is the most intensive phase. It involves both semantic and technical work.
- Vocabulary Mapping: Clinical informaticists and subject matter experts hold workshops to decide how source codes and terms will be mapped to standard OMOP concepts. This is a crucial step for ensuring clinical validity.
- ETL Development: Data engineers write the code (e.g., in SQL, Python, or using graphical ETL tools) to extract data from source systems, apply the mapping logic, transform the data structure to fit the OMOP tables, and load it into the new database. Solutions like Lifebit’s Data Factory to map to OMOP can streamline this technical process.
- Data Quality (Validation & Iteration): Your first ETL run will not be perfect. Before analyzing, rigorously validate your conversion. Run the OHDSI Data Quality Dashboard to check for conformance to the CDM standard and plausibility of the clinical data. Review the results with clinicians and data experts. This is an iterative process: find issues, fix the ETL logic, and re-run the validation until the data is reliable and meaningful.
- Engage the Community: You are not alone. The OHDSI Forums are a welcoming place to ask questions, share experiences, and learn from others who have gone through the same process.
The Future: Scaling and Integrating OMOP
The OMOP story is still being written, with a focus on scaling globally and integrating with other standards.
Global Scaling and Regional Coordination
Different regions face unique challenges, from varying data privacy regulations to different healthcare structures. This requires regional coordination. For example, Canadian organizations are working to establish a unified Canadian OHDSI chapter, sharing best practices and building a stronger national research infrastructure, as seen in events like the Pan-Canadian OMOP Common Data Model engagement event.
Bridging Clinical Care and Research with FHIR
One of the most promising future directions is integrating OMOP with Fast Healthcare Interoperability Resources (FHIR). While FHIR excels at real-time, point-to-point data exchange for clinical care (e.g., sending a lab result from the lab to the EHR), OMOP provides the optimized structure for large-scale population analytics. Combining them creates a seamless pipeline from patient care to research insights. The community is actively developing standardized mappings and open-source tools to transform FHIR resources (like Patient, Observation, and MedicationRequest) into the corresponding OMOP tables. This integration allows clinical data acquired via FHIR to be transformed into the OMOP format for robust, standardized analysis, potentially in near real-time.
AI, Machine Learning, and Federated Analytics
Standardized OMOP data is essential for training robust AI and machine learning models that can generalize across different populations. Federated analyticswhere analyses run on data at its sourceis the next frontier. At Lifebit, our federated biomedical data platform enables this secure, compliant analysis across OMOP-standardized datasets, allowing global collaboration while keeping data secure. The future is about smarter integration, more sophisticated analytics, and global collaboration that respects local governance.
Worried About OMOP Timelines and Skills? Read This Before You Start
How long does it take to convert data to the OMOP CDM?
The timeline varies significantly. A small, clean dataset might take a few months, but a large, complex system from a hospital or national registry can take six months to a year or more. A typical project breaks down into phases:
- Scoping and Assessment (1-2 months): Understanding the source data and planning the project.
- ETL Development and Mapping (3-9 months): This is the longest phase, involving iterative development of the transformation logic and clinical validation of the mappings.
- Validation and Quality Control (1-3 months): Rigorously testing the final data and iterating on fixes.
The key factors influencing the timeline are the size and complexity of your source data, its initial quality (clean, well-documented data is faster to map), and your team’s expertise and resources. It’s a significant upfront investment that pays off in long-term analytical speed and reusability.
Is OMOP only for clinical trial data?
No, this is a common misconception. OMOP was specifically designed for observational data from real-world healthcare settings. This includes electronic health records (EHRs), administrative claims data, disease registries, and patient-reported outcomes. While clinical trials are controlled environments designed to test efficacy, OMOP helps us understand treatment effectiveness and safety in routine clinical practice, open uping the power of real-world evidence.
What skills are needed for an OMOP change project?
Successful OMOP conversion requires a multidisciplinary team. You’ll need:
- Data engineers to handle the technical ETL (Extract, Transform, Load) process using SQL, Python, or R.
- Clinical informaticists to bridge the gap between the technical and clinical worlds. They lead the vocabulary mapping process, ensuring that a source code for “heart failure” is correctly mapped to the standard OMOP concept, preserving its precise clinical meaning.
- Database administrators to set up and maintain the OMOP CDM instance.
- Subject matter experts (clinicians or researchers) to validate that the transformed data accurately reflects the original clinical reality and is fit for research purposes.
- Project managers to coordinate the team and manage timelines.
The OHDSI community and partners like Lifebit provide extensive documentation, training, and support to guide organizations through this process.
Does converting to OMOP mean we lose our original data?
Absolutely not. The OMOP conversion process is non-destructive. Your original source data remains untouched in its native system. The ETL process extracts a copy of the data, transforms it, and loads it into a new, separate database that conforms to the OMOP CDM structure. This OMOP instance then serves as the standardized layer for research and analysis, while the original source systems continue to function as they always have for clinical care or administrative purposes.
Turn Every Patient Into Evidence: Standardize with OMOP and Ship AI‑Ready Insights Now
We’ve seen how the OMOP Common Data Model transforms siloed healthcare data into a single, coherent language for global research. By standardizing data structure and content, OMOP enables collaborative studies, accelerates real-world evidence generation, and open ups insights while protecting patient privacy through federated analysis.
The future of medicine grows even more exciting when OMOP is combined with artificial intelligence and machine learning. These advanced techniques thrive on the large, consistent datasets that OMOP provides. When paired with a federated infrastructure, this creates an environment for unprecedented findy.
At Lifebit, our secure federated AI platform is built to accelerate this future. Our platform components, including the Trusted Research Environment (TRE) and R.E.A.L. (Real-time Evidence & Analytics Layer), deliver real-time insights and AI-driven analytics across global data ecosystems. The future of medicine lies in learning from every patient experience, and OMOP provides the foundation.
Ready to see what your data can do when it speaks a common language? Unlock your biomedical data with a federated platform and join the global community reshaping healthcare research.

