The Multi-Omics Marvel: How Integrated Data is Transforming Disease Understanding

From Siloed Data to Saved Lives: How Multi-Omics Analytics Transforms Patient Outcomes
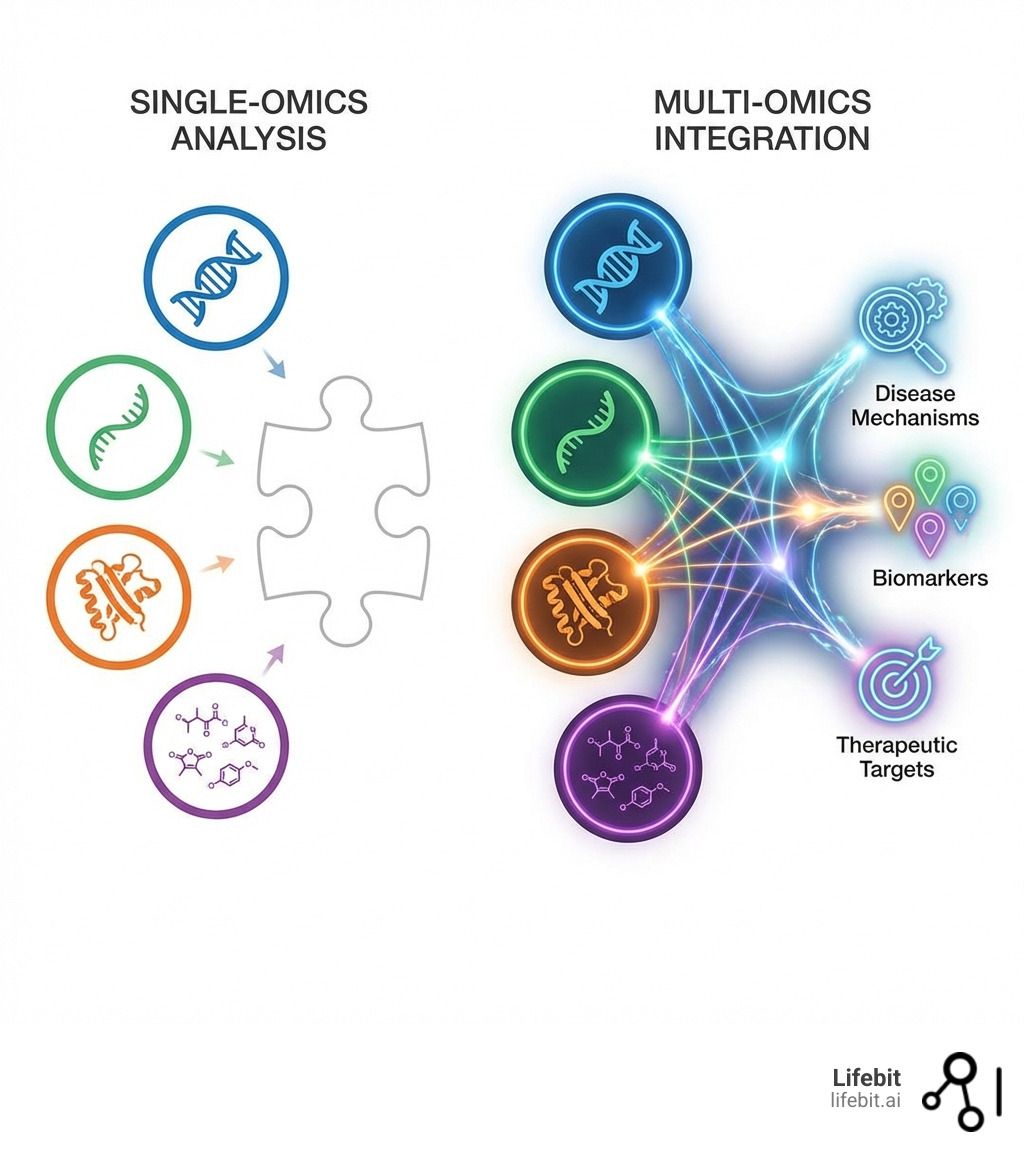
Multi-omics analytics is the integration of data from multiple biological layers—genomics, transcriptomics, proteomics, metabolomics, and more—to reveal how diseases actually work at a systems level. Here’s what it delivers:
What Multi-Omics Analytics Offers:
- Holistic Disease Understanding: Connects DNA, RNA, proteins, and metabolites to see the complete biological story
- Better Biomarker Findy: Finds disease signatures that single-layer analysis misses
- Precision Medicine: Predicts treatment response and personalizes therapies based on integrated data
- New Drug Targets: Uncovers hidden disease drivers by analyzing interactions across biological systems
- Clinical Translation: Powers real-world applications in cancer, infectious disease, and chronic conditions
The numbers tell the story of explosive growth. Multi-omics publications grew from zero in 2000 to over 1,400 per year by 2021. The second phase of the Human Microbiome Project invested $170 million specifically in integrating patient data across multiple omics datasets. Why? Because looking at just one biological layer—like genes alone—is like trying to understand a movie by reading only the script. You miss the actors, the cinematography, the music, and everything that makes it work.
Traditional single-omics approaches capture isolated snapshots: your genetic code, or your protein levels, or your metabolic state. But diseases don’t respect these boundaries. Cancer mutations change gene expression, which alters protein production, which shifts metabolism. Diabetes involves genetic risk, gut microbiome changes, metabolic dysfunction, and immune responses—all at once.
That’s where multi-omics analytics changes everything. By integrating data across biological layers, researchers can finally see the interactions that drive disease. They can move from “this gene is associated with cancer” to “here’s exactly how this mutation triggers a cascade through RNA, proteins, and metabolism to fuel tumor growth.”
The challenge? These datasets are massive, heterogeneous, and often siloed across institutions. Genomic data doesn’t look like proteomic data. Missing values are common. Batch effects distort results. And moving sensitive patient data across borders or organizations creates regulatory nightmares.
Modern multi-omics analytics solves this with federated, cloud-native platforms that analyze data where it lives—without moving it. Machine learning and AI now automate integration, find hidden patterns, and predict outcomes at scales impossible just five years ago. Over 99 software tools and 99 databases now exist specifically for multi-omics analysis, reflecting both the opportunity and the complexity.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, where we’ve spent over a decade building platforms that power multi-omics analytics for global pharma, public health agencies, and research institutions. My work spans computational biology, AI-driven genomics, and federated data systems—helping organizations integrate siloed datasets securely to accelerate precision medicine and drug findy.
Multi-omics analytics vocab explained:
- AI in Personalized Medicine
- Precision Medicine Trends You Can’t Ignore in 2025
How to Map Disease Complexity with Multi-Omics Layers
Biological systems are incredibly intricate, operating through a symphony of molecules and processes. To truly understand how life works, and how it goes awry in disease, we can’t just look at one instrument in the orchestra. We need to hear the whole performance. This is the fundamental premise of multi-omics analytics, which brings together data from various ‘omics’ layers, each providing a unique perspective on biological activity.
At the heart of cellular biology lies the central dogma: DNA makes RNA, and RNA makes protein. But this isn’t a simple linear chain; it’s a dynamic, interconnected network where feedback loops and environmental factors constantly influence the flow of information. Each ‘omic’ layer provides a distinct lens into this complex system:
Genomics: The Blueprint—But Not the Whole Story
Genomics is the study of an organism’s entire genetic material, or genome. Think of it as the master blueprint for building and maintaining an organism. Technologies like Whole Genome Sequencing (WGS) and Whole Exome Sequencing (WES) allow us to read this blueprint, identifying not just single nucleotide polymorphisms (SNPs) but also larger structural changes like copy number variations (CNVs) and insertions/deletions. Understanding these genetic variations is crucial for assessing inherited disease risk and identifying predispositions. In cancer, for example, identifying somatic mutations (changes in DNA that occur after conception) can pinpoint drivers of tumor growth. However, a static blueprint alone doesn’t tell us which genes are active and how they are regulated in response to the environment. Our Genomics expertise helps us decode this foundational layer.
Epigenomics: The Regulatory Overlay
Sitting between the static genome and the dynamic transcriptome is the epigenome. Epigenomics studies the chemical modifications to DNA and its associated proteins that regulate gene expression without altering the DNA sequence itself. Key mechanisms include DNA methylation, which can silence genes, and histone modifications, which can make genes more or less accessible for transcription. The epigenome acts as a crucial interpreter of environmental signals—like diet, stress, and toxin exposure—translating them into changes in gene activity. This layer is vital for understanding how lifestyle and environment contribute to disease, providing a mechanistic link between genetic predisposition and phenotypic outcome.
Transcriptomics: The Real-Time Instructions
If genomics is the blueprint, transcriptomics is the real-time activity log of which parts of that blueprint are being actively read and converted into RNA molecules. RNA sequencing (RNA-seq) measures gene expression levels, showing which genes are turned “on” or “off,” and to what extent. It also reveals alternative splicing events and the activity of non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which are powerful regulators of gene activity. This dynamic layer provides a snapshot of cellular function at a specific moment, offering a much more immediate view of what the cell is doing compared to its static DNA.
Proteomics & Metabolomics: The Machinery in Action
Moving further down the central dogma, proteomics is the large-scale study of proteins, the workhorses of the cell. Proteins carry out nearly all cellular functions, from catalyzing metabolic reactions to building cellular structures and transmitting signals. Mass spectrometry is a key technology here, allowing us to identify and quantify protein levels, understand their interactions, and crucially, detect post-translational modifications (PTMs). PTMs like phosphorylation or ubiquitination act as molecular switches that can dramatically alter a protein’s function, stability, or location, providing a layer of regulation that is invisible at the transcriptomic level.
Metabolomics, on the other hand, focuses on metabolites—the small molecules that are the end products of cellular processes. These include sugars, amino acids, lipids, and more. Analyzing shifts in metabolic pathways, such as the Krebs cycle or glycolysis, can reveal critical insights into cellular energy status and disease mechanisms like the Warburg effect in cancer. Metabolites reflect the immediate physiological state of a cell or organism, acting as a direct readout of biochemical activity and environmental interactions. Analyzing metabolic pathways can reveal critical insights into disease mechanisms and is a powerful approach for Biomarker Discovery.
Together, proteomics and metabolomics reveal the actual machinery in action, showing the direct consequences of genetic, epigenetic, and transcriptional changes on cellular function and health.
How to Gain a Multi-Omics Advantage for Precision Medicine
Why bother with all these layers when we could just focus on one? The simple truth is that biological systems are interconnected, and diseases are rarely caused by a single isolated factor. Relying on single-omics data is like trying to understand an elephant by only touching its trunk. You get a piece of the picture, but you miss the immense, complex whole. This is where the true power of multi-omics analytics shines.
See the Whole Disease, Not Just a Piece
Multi-omics analytics provides a holistic understanding that single-omics approaches simply cannot match. By integrating diverse datasets—genomic, transcriptomic, proteomic, metabolomic, and even microbiomic—we can map the intricate interplay between these layers. This allows us to move beyond mere correlation and start uncovering the hidden disease drivers and causal relationships. For complex diseases like cancer, diabetes, or autoimmune disorders, where multiple genetic, environmental, and lifestyle factors converge, this comprehensive view is indispensable. We can identify novel drug targets that address multiple pathways simultaneously, leading to more effective therapies, and stratify patients into more precise groups for targeted interventions.
A major limitation of classical omic studies is the isolation of only one level of biological complexity. We know that the genome provides the instructions, the transcriptome shows what instructions are being used, and the proteome and metabolome reflect the actual cellular activity. By bringing these together, we close the genotype-phenotype gap, directly linking genetic predispositions to observable traits and disease states.
Predict and Personalize: The Power for Precision Medicine
The ultimate goal of much of this research is to advance Precision Medicine. With multi-omics analytics, we can predict treatment response with greater accuracy, assess individual disease risk more comprehensively, and develop truly custom therapies. Imagine predicting whether a cancer patient will respond to a specific chemotherapy based on their unique genomic, proteomic, and metabolic profile, rather than relying on broad population averages. Or identifying individuals at high risk for developing a chronic condition years in advance, allowing for preventative measures.
This is achieved by identifying multi-omics biomarker signatures—patterns across different data layers that are indicative of a particular disease state, prognosis, or response to treatment. These integrated signatures are far more robust and informative than single-omics biomarkers, offering a powerful tool for early diagnosis, monitoring disease progression, and guiding therapeutic decisions.
How the Multi-Omics Findy Engine Actually Works
So, how do we actually weave these disparate biological threads into a coherent mix of understanding? The process of multi-omics analytics is a sophisticated blend of data science, bioinformatics, and biological interpretation. It starts with the raw data and moves through rigorous steps of processing and integration to extract meaningful insights.
Data generation techniques, such as next-generation sequencing, mass spectrometry, and microarrays, produce vast amounts of raw data. This data then undergoes pre-processing, which involves cleaning, filtering, and quality control to remove noise and ensure reliability. Normalization steps are crucial to make data comparable across different experiments and samples. A particularly tricky aspect is batch effect correction, which accounts for technical variations introduced during sample preparation or measurement across different batches, preventing them from masquerading as biological signals.
Step-by-Step: The Multi-Omics Analysis Workflow
The journey from raw data to biological findy in multi-omics analytics typically follows several key stages:
- Data Acquisition: Collecting data from various omics platforms (genomics, transcriptomics, proteomics, etc.).
- Quality Control & Pre-processing: Ensuring data integrity, removing artifacts, and standardizing formats.
- Feature Selection: Identifying the most relevant genes, proteins, or metabolites within each omics layer that are most likely to contribute to the biological question at hand.
- Data Integration: This is the core of multi-omics analytics. It involves combining the processed data from different layers. There are several common integration methods:
- Canonical Correlation Analysis (CCA) and its variants: These statistical methods identify linear relationships between two sets of variables, finding latent variables that maximize correlation across omics types. Advanced versions like Sparse CCA and Regularized Generalized CCA (RGCCA) are used for high-dimensional data.
- Partial Least Squares (PLS) and its extensions: PLS methods are particularly useful for predicting a response variable from multiple omics datasets or for finding components that explain variance across datasets.
- Matrix Factorization methods: Techniques like Joint and Individual Variation Explained (JIVE) or Non-negative Matrix Factorization (NMF) decompose multi-omics data into shared and unique components, allowing us to identify common patterns and layer-specific insights.
- Probabilistic-based Methods: Models like iCluster or Multi-Omics Factor Analysis (MOFA) use statistical frameworks to infer latent variables that capture underlying biological processes driving variation across omics datasets.
- Kernel-based Methods: These methods use kernel functions to capture non-linear relationships between variables, such as Multiple Kernel Learning (MKL).
- Network-based Methods: Approaches like Similarity Network Fusion (SNF) build patient similarity networks from each omics type and then fuse them into a single, more robust network, often used for disease subtyping.
- Deep Learning Approaches: As we’ll discuss, these advanced methods are increasingly used for complex, non-linear integration.
- Biological Interpretation & Pathway Analysis: Once integrated, the data is analyzed to identify enriched biological pathways, functional modules, and key regulatory networks. This helps translate statistical findings back into biological meaning.
Breaking Barriers: Challenges in Multi-Omics Analytics
While the promise of multi-omics analytics is immense, the path is not without its problems. We grapple with several significant challenges:
- Data Heterogeneity: Each omics technology produces data with unique characteristics, noise profiles, and scales. Integrating these vastly different data types is like trying to combine apples, oranges, and zebras – each requires a different approach.
- High Dimensionality: Multi-omics datasets are notoriously high-dimensional, often containing thousands to millions of features (genes, proteins, metabolites) for a relatively small number of samples. This “curse of dimensionality” can lead to overfitting and computational bottlenecks.
- Missing Data: It’s common for individual omics datasets to have missing values due to technical limitations or experimental design. Robust integration methods must be able to handle this incompleteness effectively.
- Computational Complexity: Processing, integrating, and analyzing these massive datasets requires significant computational resources and sophisticated algorithms.
- Data Security and Governance: Dealing with sensitive patient data across multiple institutions, especially across different countries with varying regulations, presents immense challenges in data privacy, security, and ethical use. This is where solutions like Lifebit’s secure, federated data analysis platforms come into play, enabling compliant research without moving raw data. For More info about secure, federated data analysis, you can dig into our approach.
To overcome these, we need innovative solutions, both computational and infrastructural.
The AI Revolution: How Machine Learning Boosts Multi-Omics
The sheer scale and complexity of multi-omics analytics demand powerful computational tools. This is where the AI revolution, particularly machine learning and deep learning, has emerged as a game-changer. Machine learning is synergistically used with multi-omics data integration, playing a pivotal role in the findy of novel biomarkers through various statistical frameworks and sophisticated latent variable models.
Deep learning, a subset of machine learning, offers a powerful way to model complex, non-linear relationships within and between omics datasets. Techniques like autoencoders, variational autoencoders (VAEs), and generative adversarial networks (GANs) are proving invaluable for:
- Factor Analysis and Dimensionality Reduction: Identifying latent biological factors that explain variation across multiple omics layers.
- Predictive Modeling: Building robust models to predict disease outcomes, drug responses, or patient stratification.
- Pattern Recognition: Uncovering subtle, intricate patterns in the data that human analysts might miss.
- Data Imputation and Augmentation: Filling in missing data and generating synthetic data to improve training sets.
- Batch Effect Correction: Attenuating technical variations to ensure biological signals are accurately captured.
For a deeper dive, Scientific research on AI in multi-omics extensively covers these advancements. From our perspective, AI for Genomics is rapidly accelerating our ability to derive insights from vast genomic datasets, especially when combined with other omics.
More recently, foundation models, initially popularized in natural language processing (NLP), are being adapted for multi-omics analytics. These models are pre-trained on vast, diverse biological datasets and can then be fine-tuned for specific tasks, offering unprecedented generalization capabilities. This approach is particularly promising for addressing data scarcity in certain research areas, by leveraging knowledge gained from broader biological contexts.
The Analyst’s Toolkit: How Lifebit Empowers Multi-Omics Analytics
At Lifebit, we understand that open uping the full potential of multi-omics analytics requires more than just powerful algorithms; it demands an infrastructure that is secure, scalable, and user-friendly. Our platform is designed to be the ultimate Data Analytics Platform for complex biomedical data.
We provide a next-generation federated AI platform that enables secure, real-time access to global biomedical and multi-omic data. With built-in capabilities for harmonization, advanced AI/ML analytics, and federated governance, we power large-scale, compliant research and pharmacovigilance across biopharma, governments, and public health agencies.
Our web-based and cloud-native solutions offer:
- Seamless Data Integration: Our Data Integration Platform is built to handle the heterogeneity and scale of multi-omics data, connecting disparate datasets into a unified view.
- End-to-End Workflow Automation: From raw data processing to advanced statistical modeling and biological interpretation, our platform streamlines the entire multi-omics analytics workflow, reducing manual effort and accelerating findy.
- Secure, Federated Analysis for Global Collaboration: Our Federated Data Platform leverages Trusted Research Environments to allow researchers to analyze sensitive data where it resides, without compromising privacy or regulatory compliance. This is critical for international collaborations involving patient data from the UK, USA, Canada, Europe, and beyond. Our Data Governance Platform ensures that all analyses adhere to the strictest ethical and legal standards.
Our platform components, such as the Trusted Data Lakehouse (TDL) and R.E.A.L. (Real-time Evidence & Analytics Layer), deliver real-time insights, AI-driven safety surveillance, and secure collaboration across hybrid data ecosystems, making complex multi-omics analytics accessible and actionable for our global users.
How Multi-Omics Analytics is Saving Lives in the Real World
The theoretical advantages of multi-omics analytics are compelling, but its true value is measured in its real-world impact. Across various domains of health and disease, integrated omics data is providing unprecedented insights, leading to breakthroughs that were previously unimaginable.
Cancer: Unmasking Tumor Secrets for Targeted Therapies
Cancer is a prime example where multi-omics analytics is revolutionizing our understanding and treatment approaches. Large-scale consortium projects, such as The Cancer Genome Atlas (TCGA) and the Pan-Cancer Analysis of Whole Genomes (PCAWG), have generated immense multi-omics datasets across thousands of tumors and dozens of cancer types. By integrating genomic mutations, epigenomic marks, gene expression profiles, protein levels, and metabolic changes, researchers are gaining a comprehensive view of tumor heterogeneity—the diverse cell populations within a single tumor. This has enabled:
- Identifying Driver Mutations: Pinpointing the specific genetic alterations that initiate and sustain cancer growth.
- Understanding Resistance Mechanisms: Elucidating why some cancers become resistant to therapies, paving the way for new treatment strategies.
- Molecular Subtyping: Classifying cancers into more precise subtypes, which can significantly improve prognosis and guide targeted therapies. For example, in glioblastoma, multi-omics integration has helped classify tumors into subtypes (e.g., proneural, mesenchymal) that respond differently to treatment, a distinction not always clear from histology alone. Similarly, analysis of triple-negative breast cancer has revealed that vulnerabilities in DNA repair pathways can be targeted with specific drugs like PARP inhibitors, but only in tumors with the right molecular signature across multiple omic layers.
Integrated analytics accelerates findy, moving us closer to truly personalized cancer treatments. Our AI-Powered Biomarker Discovery capabilities are particularly impactful in this area.
Infectious Disease & Microbiome: Outpacing Outbreaks and Chronic Illness
Multi-omics analytics is also proving critical in the fight against infectious diseases and in understanding the complex role of the microbiome in health and disease.
- Host-Pathogen Interactions: By integrating host omics data (e.g., immune response genes, proteins) with pathogen omics data (e.g., bacterial genomes), we can understand how pathogens interact with the host at a molecular level. This is crucial for developing new diagnostics and treatments.
- COVID-19 Research: During the pandemic, multi-omics analytics helped researchers understand the distinct serum profiles associated with severe COVID-19 in pregnancy, including greater complement activation and dysregulation of serum lipids.
- Human Microbiome Project: The second phase of the $170 million Human Microbiome Project was specifically focused on integrating patient data to different omic datasets, revealing insights into conditions like inflammatory bowel disease and preterm birth. This research has shown how the metabolic output of gut bacteria (the ‘metabolome’) can directly influence the host’s immune system (via transcriptomics and proteomics) and even contribute to the efficacy of cancer immunotherapies. This project, among others, has profoundly advanced our understanding of the gut microbiome’s role in both local and systemic chronic illnesses.
- Vaccine Development (Systems Vaccinology): By combining omics data from individuals before and after vaccination, systems vaccinology uses multi-omics analytics to understand the immune response, predict vaccine efficacy, and identify potential side effects. This helps us develop more effective and safer vaccines.
Neurodegenerative and Chronic Diseases: Decoding Complexity Over Time
The power of multi-omics extends far beyond cancer and infectious disease, offering profound insights into chronic and neurodegenerative conditions like Alzheimer’s disease, Parkinson’s disease, and diabetes. These diseases often develop over decades, driven by a complex interplay of genetic risk, environmental factors, and aging.
In Alzheimer’s disease, for instance, a single-omics approach falls short. Genomics identifies risk factors like the APOE4 allele, but can’t predict who will develop the disease or when. Multi-omics analytics connects the dots: integrating genomics with epigenomics reveals how environmental factors might alter the expression of risk genes. Transcriptomics of brain tissue or even blood cells can identify inflammatory pathways that become active years before symptoms appear. Proteomics quantifies the buildup of amyloid-beta and tau proteins, the pathological hallmarks of the disease, while metabolomics can detect early changes in brain energy metabolism or lipid profiles. By combining these layers, researchers can build predictive models for early diagnosis, stratify patients for clinical trials, and identify novel therapeutic targets aimed at halting the disease process rather than just managing symptoms.
The Next Frontier: Single-Cell and Spatial Multi-Omics
Perhaps one of the most exciting advancements in multi-omics analytics is the emergence of single-cell and spatial multi-omics. Traditional omics studies typically analyze bulk tissue samples, which average out the signals from millions of cells. This can mask crucial information about cellular heterogeneity—the differences between individual cells—which is vital for understanding disease progression and treatment response.
- Single-Cell Multi-Omics: This technology allows us to perform multiple omics measurements (e.g., genomics, transcriptomics, epigenomics) from individual cells. This unprecedented resolution helps us understand how individual cells contribute to disease, identify rare cell populations, and track cellular transitions in health and disease. For instance, single-cell multi-omics is used to integrate transcriptomes with methylomes or to query epigenomes using techniques like single-cell ATAC-seq.
- Spatial Multi-Omics: Complementing single-cell approaches, spatial omics assays tissues while preserving the relative spatial orientation of cells. This means we can create detailed maps of complex tissues like tumors, the brain, or developing organs. For example, in cancer research, spatial multi-omics can reveal how cancer cells interact with surrounding immune cells and stromal cells in the tumor microenvironment. We can see which signaling pathways are active at the invasive front of a tumor versus its core, providing critical clues for understanding metastasis and designing therapies that disrupt these cellular neighborhoods. This is changing our understanding of tissue architecture and developmental biology. Our resource on Single-cell and Spatial methods provides more details on this burgeoning field.
These cutting-edge approaches are pushing the boundaries of biological findy, revealing secrets hidden within the microscopic world and paving the way for new diagnostic and therapeutic strategies.
How to Prepare for the Future of Health with Integrated Multi-Omics
The journey from fragmented biological data to a full, integrated picture of health and disease has been transformative. Multi-omics analytics is no longer a niche academic pursuit; it is a fundamental approach that is redefining how we conduct biological research, develop medicines, and ultimately improve patient care.
The benefits are clear: a more comprehensive understanding of biological systems, the findy of robust biomarkers, and the ability to deliver truly personalized medicine. We’ve seen its power in deciphering the complexities of cancer, outpacing infectious diseases, and solving the mysteries of our microbiome. With the advent of single-cell and spatial multi-omics, we are now probing biological systems at an unprecedented resolution.
Looking ahead, the future of multi-omics analytics is inextricably linked with AI-driven findy. Machine learning and deep learning models will continue to evolve, offering even more sophisticated ways to integrate, interpret, and predict from these vast datasets. The application of foundation models, initially developed for other complex data types, promises to open up new levels of insight and accelerate breakthroughs across the globe, including in the UK, USA, Canada, and Europe.
However, the immense volume and sensitivity of multi-omics data necessitate robust infrastructure. The critical role of federated analysis cannot be overstated. By enabling secure, compliant analysis of data where it lives, without moving it, we can overcome data silos and accelerate global research collaborations. This approach protects patient privacy while fostering a collaborative environment essential for rapid scientific progress.
At Lifebit, we are proud to be at the forefront of this revolution, powering precision medicine globally. Our federated AI platform is designed to meet these challenges head-on, providing the secure, scalable, and intelligent solutions needed to harness the full potential of multi-omics analytics.
Don’t be left behind. The future of health is integrated, intelligent, and here. Explore how federated technology powers precision medicine and join us in building a healthier future for everyone.

