The No-Nonsense Guide to Nextflow HPC Singularity Setup

Why Nextflow HPC Singularity Integration Matters for Research at Scale
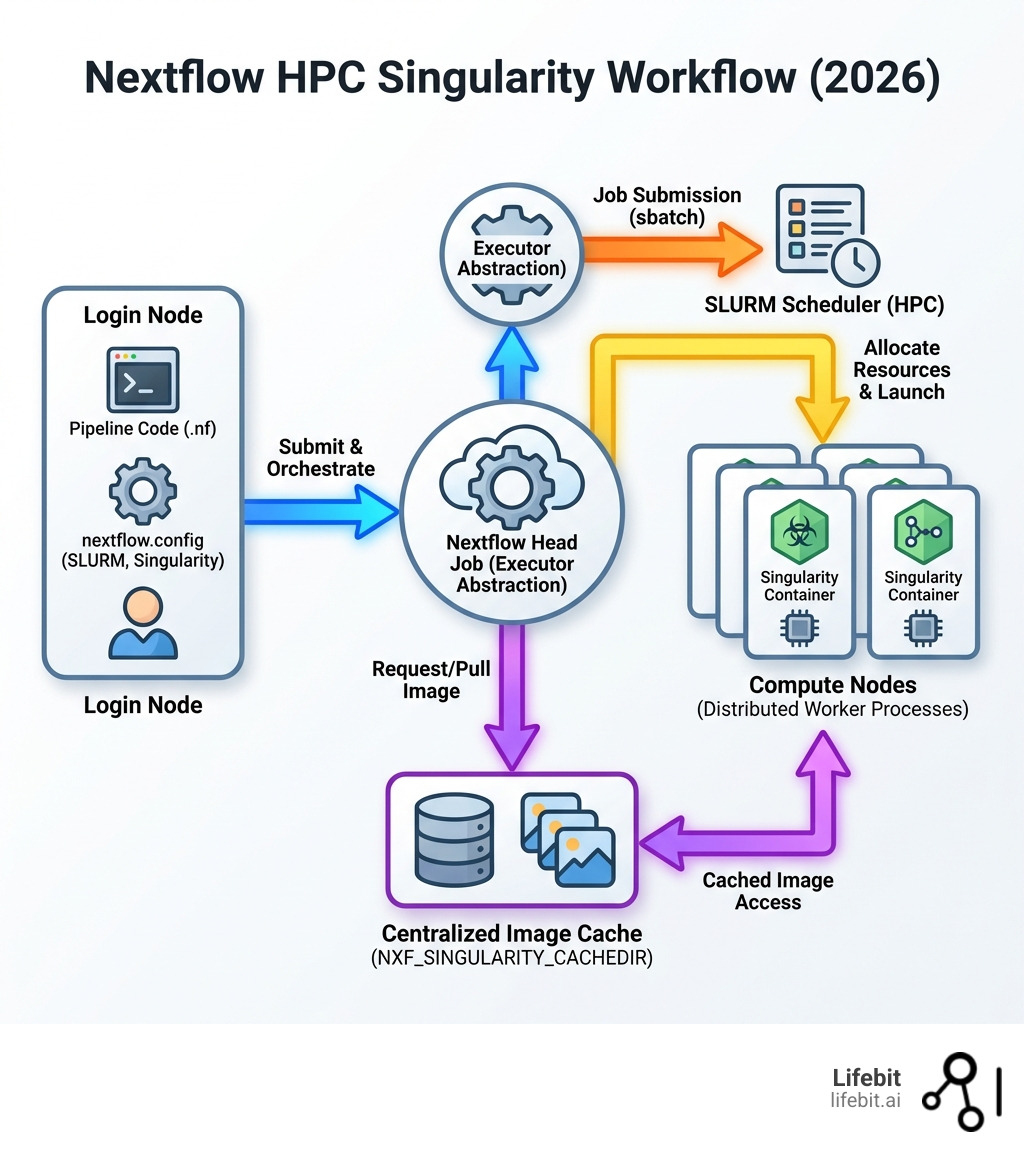
Nextflow hpcsing is the combination of Nextflow’s workflow orchestration with Singularity containers on HPC clusters managed by SLURM schedulers. This technical synergy is not merely a convenience; it is the foundation of modern computational biology and data science. When we talk about “nextflow hpcsing,” we are describing a three-tier architecture: the Nextflow engine (the brain), the SLURM scheduler (the muscle), and Singularity containers (the environment).
To configure this stack correctly, researchers must follow a rigorous setup process:
- Set the SLURM executor in
nextflow.configwithprocess.executor = 'slurm'. This tells Nextflow to stop acting as a local process manager and start acting as a job submitter. - Enable Singularity by adding
singularity.enabled = trueto your config. This ensures that every task is wrapped in a container execution command. - Configure cache directory using
NXF_SINGULARITY_CACHEDIR. Without this, Nextflow will download the same multi-gigabyte images into every work directory, quickly exhausting storage quotas. - Specify resource limits with directives like
cpus,memory,queue, andtime. These must align with your HPC’s partition limits to avoid “Job Rejected” errors. - Launch from login nodes where
sbatchcommands are available, or better yet, submit the Nextflow “head” process itself as a SLURM job to ensure the controller isn’t killed during long-running workflows. - Use process selectors (
withName,withLabel) for granular resource allocation, ensuring that a simple text-parsing script doesn’t reserve a 1TB RAM node.
The challenge most research teams face isn’t writing pipelines—it’s making them run reliably across different compute environments without rewriting code. When you’re processing terabytes of genomic data or running AI models on patient cohorts, you need a system that abstracts away infrastructure complexity while maintaining security and reproducibility. Nextflow’s dataflow programming model allows for asynchronous execution, meaning tasks run as soon as their inputs are ready, maximizing cluster utilization.
Nextflow solves this through its executor abstraction layer, which lets you run the same pipeline on your laptop, a university cluster, or cloud infrastructure by changing a single configuration line. Singularity complements this by providing rootless containerization that works on shared HPC systems where Docker’s privileged access is a non-starter. In an HPC environment, security is paramount; Singularity allows users to run containers as themselves, respecting the existing file system permissions and UID/GID mappings of the host cluster.
But configuration mistakes cost you time and money. Setting the wrong cache directory fills up home folders. Misconfigured SLURM options lead to jobs stuck in pending queues. Without proper resource allocation, you either waste compute credits or crash jobs mid-analysis. Furthermore, understanding the interaction between the Nextflow “head” process and the “worker” jobs is critical. The head process monitors the status of all submitted jobs; if the head process dies, the entire workflow stalls, even if the worker jobs are still running on the compute nodes.
As Maria Chatzou Dunford, co-founder of Lifebit and a core contributor to Nextflow, I’ve spent over a decade building computational biology tools for precision medicine and federated data analysis. I’ve helped organizations deploy nextflow hpc_sing setups that process millions of patient records across secure, compliant HPC environments. Let me show you the exact configuration steps that work.
Why Nextflow and Singularity are the Gold Standard for HPC
In scientific computing, the “it works on my machine” excuse is a death sentence for reproducibility. When we deal with multi-omic data at scale, we need a stack that is portable, scalable, and secure. This is where nextflow hpc_sing becomes the industry standard. The integration of these tools addresses the “dependency hell” that has plagued bioinformatics for decades, where conflicting versions of Python, R, or C++ libraries made it nearly impossible to replicate results across different institutions.
Nextflow provides the orchestration logic, while Singularity (now often referred to as Apptainer) handles the software environment. Together, they ensure that a pipeline developed in a London lab runs exactly the same way on an HPC cluster in New York or a federated TRE in Canada. This portability is achieved by encapsulating the entire software stack—OS, libraries, and binaries—into a single immutable file (the .sif image).
Executors are the secret sauce here. They act as an abstraction layer between your pipeline script and the underlying hardware. For a deeper dive into how this fits into the broader AI landscape, check out our AI Nextflow Complete Guide. By decoupling the workflow logic from the execution environment, Nextflow allows developers to focus on the science while the engine handles the logistics of job submission, file staging, and error recovery.
The Power of Executor Abstraction
The beauty of Nextflow lies in its “write-once-run-anywhere” philosophy. By default, Nextflow uses a local executor, running tasks as processes on the machine where it was launched. However, for real-world research, you need the power of an HPC scheduler like SLURM. SLURM (Simple Linux Utility for Resource Management) is the gatekeeper of the cluster, deciding which jobs run on which nodes based on priority and resource availability.
When you switch the executor to SLURM, Nextflow stops running tasks locally. Instead, it automatically generates sbatch scripts for every single task in your pipeline and submits them to the cluster queue. It then supervises these tasks, handling retries and resource monitoring without you ever having to manually write a submission script. This is particularly powerful for pipelines with thousands of tasks; manually managing that many sbatch scripts would be an administrative nightmare and prone to human error.
Why Singularity Beats Docker on Clusters
While Docker is the king of the cloud, it is rarely allowed on shared HPC clusters. Why? Because Docker requires root privileges (the “sudo” problem), which is a massive security risk in a multi-user environment. If a user has root access inside a Docker container, they can potentially escalate those privileges to the host system, compromising the entire cluster.
Singularity was built specifically for HPC. It allows for rootless execution, meaning you can run containers with your own user permissions. It also integrates seamlessly with shared file systems (like Lustre, GPFS, or Isilon), which is essential for reading massive datasets. Unlike Docker, which uses a complex layered file system, Singularity uses a single-file image format that is much easier to manage on network-attached storage. By using nf-core Standardised Bioinformatics Pipelines, you ensure that every tool in your workflow is pre-packaged and ready to go without worrying about dependency hell. This standardization is what allows large-scale consortia to collaborate effectively, knowing that their results are comparable across different compute sites.
Configuring the SLURM Executor for nextflow hpc_sing
To get started, you need to tell Nextflow how to talk to your cluster. This is done in the nextflow.config file, which acts as the central nervous system for your workflow’s execution parameters. A well-configured config file can mean the difference between a pipeline that finishes in hours and one that hangs for days.
The most basic configuration requires setting the process.executor to slurm. However, production-grade environments require more nuance. Here is a snippet of what a production-ready config looks like, including settings for job throttling and resource defaults:
process {
executor = 'slurm'
queue = 'compute'
cpus = 1
memory = '8 GB'
time = '4h'
clusterOptions = '--account=my_project_id'
}
executor {
queueSize = 50
submitRateLimit = '10 sec'
pollInterval = '30 sec'
exitReadTimeout = '10 min'
}
The queueSize directive is crucial—it prevents you from overwhelming the SLURM controller by limiting how many jobs Nextflow can have in the queue at once. If you submit 5,000 jobs simultaneously, the cluster’s management node might slow down for all users, leading to a stern email from your sysadmin. The submitRateLimit further protects the scheduler by spacing out the sbatch commands. Some large clusters, like NYU HPC, support a queueSize as high as 1900, but always check with your sysadmin first to understand the specific limits of your environment.
Essential Environment Variables for nextflow hpc_sing
One of the biggest mistakes we see is users letting Nextflow download Singularity images into their home directory. Home directories on HPC are usually small (often limited to 20-50 GB) and will hit their quota in minutes when downloading large genomic toolsets. Furthermore, home directories are often on slower storage tiers, which can bottleneck the container startup process.
You must set the NXF_SINGULARITY_CACHEDIR environment variable. This tells Nextflow to store the .sif image files in a centralized, high-capacity storage area, such as a scratch or project directory. We recommend adding this to your .bashrc or your job submission script:
export NXF_SINGULARITY_CACHEDIR=/path/to/large/scratch/nxf_images
This ensures that nf-core Nextflow Pipelines only download a tool container once. If multiple users point to the same directory, they can share the images, saving time, bandwidth, and storage space for everyone on your team. This is a critical best practice for institutional HPC setups.
Launching Workflows: Login Nodes vs. SLURM Jobs
Where you run the nextflow run command matters significantly for the stability of your analysis.
| Method | Description | Best For |
|---|---|---|
| Login Node | Run directly on the terminal. | Short test runs (not recommended for production). |
Interactive (salloc) |
Request a single node for interactive use. | Debugging, setup, and small-scale testing. |
| Wrapper Batch Job | Submit the Nextflow “head” process as a SLURM job. | Production runs and long-running pipelines. |
Running on a login node for days is risky. Login nodes are shared resources intended for light tasks like code editing and job submission. If your Nextflow process consumes too much memory or CPU while managing thousands of sub-jobs, the system’s “OOM Killer” or a watchdog script will terminate it. The safest way is to create a small “wrapper” script (e.g., run_pipeline.sh) and submit it with sbatch. This ensures the Nextflow controller process itself is protected, has its own dedicated resources, and will continue running even if you disconnect from the cluster.
Mastering Singularity Containers and Centralized Caching
Once the executor is set, you need to enable the Singularity engine. This is the component that ensures your software environment is identical every time the pipeline runs. In your nextflow.config, you should define a singularity scope to manage how containers are handled.
singularity {
enabled = true
autoMounts = true
runOptions = '--bind /scratch,/project/data'
cacheDir = '/path/to/shared/containers'
pullTimeout = '20 min'
}
The autoMounts setting is a lifesaver—it automatically maps your cluster’s file system paths into the container so the tools can actually “see” your data. Without this, a tool inside the container might try to read /home/user/data.fastq and fail because it only sees its own internal file system. The runOptions allow you to pass specific flags to the singularity run command, such as --nv for GPU support or specific --bind paths for non-standard directory structures. This follows the best practices outlined in the Oregon State University documentation for scientific computing.
Setting Up a Global Image Cache
If you are working in a large department, don’t have every researcher download their own copy of the same 5GB container. Set up a centralized directory like /cluster/data/containers and point everyone’s NXF_SINGULARITY_CACHEDIR there. This not only saves terabytes of space but also speeds up job startup times significantly. When a job starts, Singularity doesn’t have to pull an image from the internet; it simply performs a local read of the .sif file. In environments with restricted internet access (air-gapped clusters), this centralized cache is the only way to run containerized workflows.
Handling Process-Specific Resources in nextflow hpc_sing
Not all tasks are created equal. A “mapping” task might need 16 CPUs and 64GB of RAM, while a “summary” task needs only one CPU and 2GB of RAM. Nextflow allows you to handle this gracefully using withLabel or withName selectors. This prevents “resource over-provisioning,” where you request more than you need, and “under-provisioning,” where your job crashes due to lack of memory.
process {
// Default resources for all processes
cpus = 1
memory = '4 GB'
withLabel: 'big_mem' {
cpus = 16
memory = '128 GB'
time = '24h'
}
withName: 'FASTQC' {
cpus = 2
memory = '8 GB'
}
withName: 'BWA_MEM' {
cpus = 8
memory = { 32.GB * task.attempt }
errorStrategy = 'retry'
}
}
In the example above, the BWA_MEM process uses a dynamic memory allocation. If the job fails, Nextflow will retry it and double the memory for the next attempt. This is a sophisticated way to handle unpredictable data sizes. For those looking to push performance further, you can Accelerate Nextflow with our nf-copilot Feature, which helps optimize these allocations based on real-time data and historical performance metrics.
Optimizing nextflow.config for High-Throughput Performance
When running thousands of jobs, small inefficiencies lead to massive delays. Optimization in a nextflow hpc_sing environment requires a deep understanding of both the workflow’s needs and the cluster’s architecture. One of the most common bottlenecks is the metadata overhead of the file system. Every time Nextflow checks if a file exists or creates a new task directory, it performs I/O operations that can slow down the entire cluster if not managed correctly.
HPC clusters often have hard limits. For example, the CQLS cluster recommends a max_cpus of 96 and max_memory of ‘384 GB’ for a single node. If your pipeline tries to request more than the cluster’s physical limits, SLURM will reject the job instantly, and Nextflow will stop the entire workflow. Always define these “ceiling” parameters in your config to prevent crashes and ensure your pipeline is portable to smaller clusters.
Strategies for I/O and Error Handling
I/O is often the primary bottleneck in nextflow hpc_sing setups. Whenever possible, use $TMPDIR. Most HPC nodes have local SSDs (often called “node-local scratch”) that are much faster than the network-attached storage (NAS) where your main data lives. Writing intermediate files to the NAS creates network congestion; writing them to local scratch keeps the traffic local to the node.
By setting scratch = true in your process definition, Nextflow will run the task in the node’s local temporary space and only copy the final results back to the main work directory when the task is finished. This can reduce network congestion by up to 70% and significantly speed up tasks that involve many small read/write operations.
Additionally, use a robust error strategy to handle the transient failures common in HPC environments (e.g., a node going down or a temporary file system glitch):
process {
errorStrategy = { task.exitStatus in [143,137,104,134,139,140] ? 'retry' : 'finish' }
maxRetries = 3
maxErrors = '-1'
}
This tells Nextflow: “If the job was killed because it ran out of memory (Error 137) or exceeded its time limit (Error 140), don’t give up—just try again.” You can even combine this with the dynamic resource scaling mentioned earlier. For more on AI-driven optimization and how to predict the resource needs of your tasks before they run, see AI for Nextflow.
Creating Environment-Specific Profiles
Profiles allow you to switch between different setups (e.g., test, standard, nyu_hpc, aws_batch) using a simple flag: nextflow run main.nf -profile nyu_hpc. This is the key to making your pipelines truly portable. A profile can encapsulate all the SLURM and Singularity settings specific to a particular institution.
The NYU HPC profile, for instance, might set massive resource limits (up to 3000 GB of memory) and specific queue requirements for GPU nodes. By modularizing your config into profiles, you make your pipeline usable by different departments or collaborators without them having to touch the core logic of the workflow. This separation of concerns is a hallmark of professional-grade pipeline development.
Frequently Asked Questions about nextflow hpc_sing
How do I prevent Singularity from filling up my home directory?
As mentioned, always set NXF_SINGULARITY_CACHEDIR to a scratch or project directory. If you’ve already filled your home, look for a hidden folder named .singularity in your home directory. This is where Singularity stores its own internal metadata and temporary layers. You can move this folder to a larger disk and create a symbolic link, or set the SINGULARITY_TMPDIR and SINGULARITY_CACHEDIR environment variables in your shell.
Why is my Nextflow job stuck in the SLURM “PENDING” state?
This usually happens for three reasons:
- Resource Request too high: You asked for more CPUs or RAM than any single node possesses. Check the cluster’s node specifications using
sinfo -Ne. - Invalid Queue: The queue (partition) you specified doesn’t exist or you don’t have permission to use it. Ensure your
process.queuematches a valid partition. - Cluster Congestion: The cluster is simply full. Use
squeue -uto see the “REASON” column provided by SLURM. Common reasons includePriority,Resources, orAssociationJobLimit.
Can I run nf-core pipelines like ampliseq using this setup?
Absolutely. Most nf-core pipelines, including ampliseq, rnaseq, and sarek, come with a built-in -profile singularity that handles most of the heavy lifting. You just need to provide the cluster-specific SLURM details (like the partition name and account ID) in a custom config file and pass it with the -c flag.
How do I handle “Singularity pull” timeouts?
On some clusters, the internet connection on the login node is slow, causing Nextflow to time out while downloading large containers. You can increase the timeout limit by setting singularity.pullTimeout = '30 min' in your config. Alternatively, you can manually pull the image using singularity pull before running the pipeline.
What is the difference between Singularity and Apptainer?
Apptainer is the successor to Singularity under the Linux Foundation. For most Nextflow users, they are functionally identical. Nextflow’s singularity configuration block works for both. If your cluster has apptainer installed instead of singularity, Nextflow will automatically detect and use it.
Conclusion
Setting up nextflow hpc_sing is the single most important step you can take toward scalable, reproducible research. By mastering the SLURM executor, configuring centralized Singularity caching, and utilizing granular resource selectors, you transform your HPC cluster from a source of frustration into a powerful engine for discovery. The combination of Nextflow’s orchestration and Singularity’s containerization provides a robust framework that can handle the most demanding computational tasks in modern science.
However, the technical setup is only half the battle. As datasets grow into the petabyte scale and research becomes increasingly collaborative, the need for managed platforms that automate these configurations becomes clear. Managing your own nextflow.config files across multiple clusters is manageable for a single project, but for an entire organization, it requires a more centralized approach to governance, security, and cost control.
At Lifebit, we believe that your infrastructure should never be a barrier to science. Our federated AI platform is built on these very principles, enabling secure access to multi-omic data across hybrid ecosystems while maintaining the highest standards of governance and compliance. We automate the complexities of Nextflow and Singularity integration, allowing researchers to focus on generating insights rather than debugging scheduler scripts. Whether you are working in biopharma, academia, or public health, we are here to help you turn complex data into real-time insights.
Accelerate your Nextflow workflows with Lifebit and see how our TRE and Trusted Data Lakehouse can power your next breakthrough. By leveraging our expertise in federated analysis and high-performance computing, you can ensure that your research is not only fast and scalable but also fully reproducible and secure.

