The Federated Frontier: Exploring Research Environments of Tomorrow

Stop Letting Data Silos Kill Your Research Speed (And Budget)
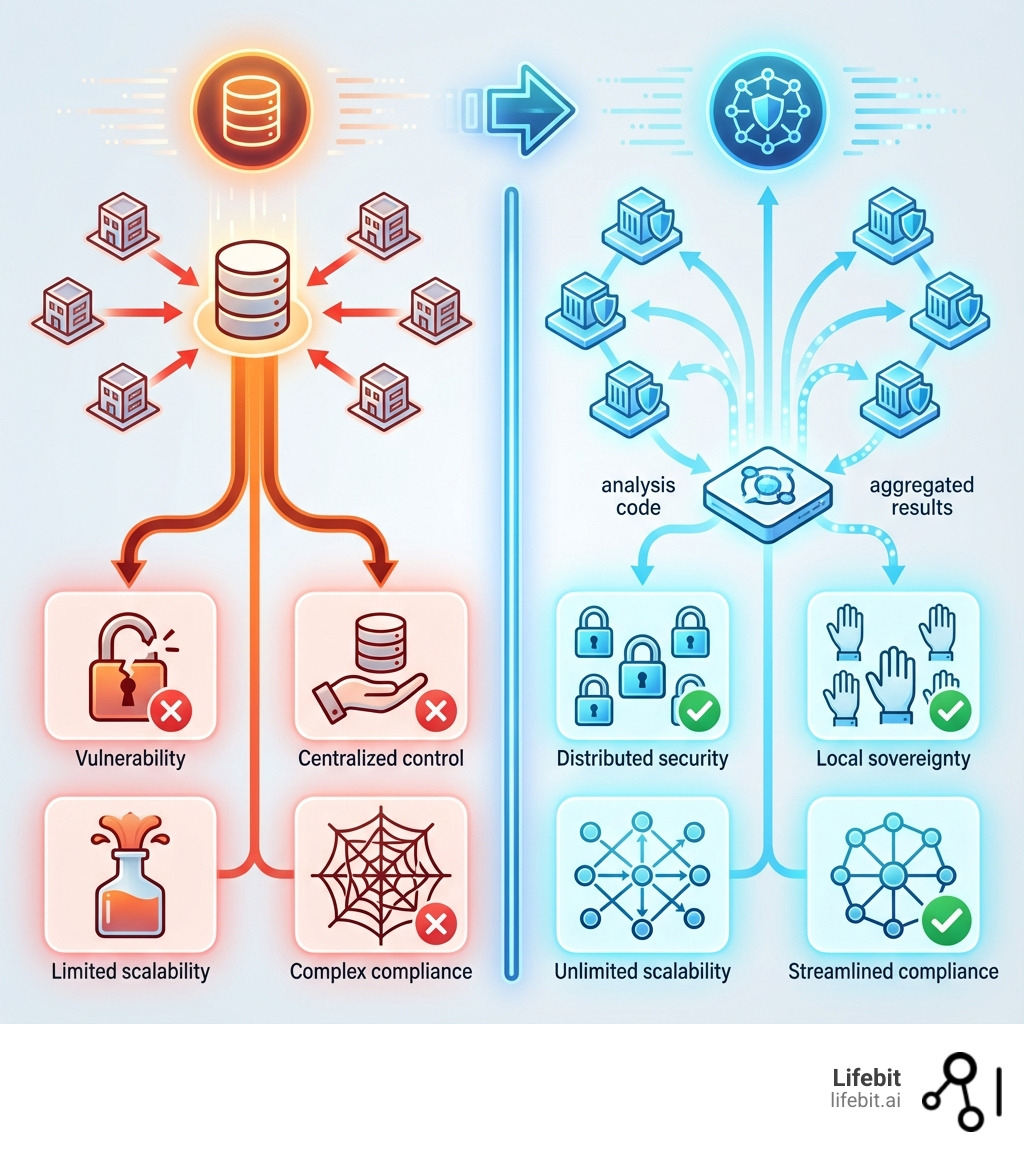
A federated research environment is a secure framework that lets researchers analyze sensitive data across multiple institutions without moving it. This model brings computation to the data, not the other way around.
Key characteristics of federated research environments:
- Data stays in place at the source, maintaining sovereignty and control.
- Analysis code travels to each data location via standardized APIs.
- Aggregated results are returned securely, protecting patient-level data.
- Centralized metadata enables findy, while raw data stays distributed.
- Access is controlled through strictly governed Trusted Research Environments (TREs).
Scientific data management faces a critical crisis. Data volumes are exploding, results are fragmented, and a severe reproducibility crisis threatens modern research.
Traditional centralized data warehouses cannot scale. Moving petabytes of genomic and biomedical data is prohibitively expensive, fragmentation hampers collaboration, and data sovereignty concerns make centralization politically untenable.
The solution is federation. Instead of centralizing data, federated environments enable analysis where data lives. Organizations collaborate through shared standards, each retaining control over its data while enabling secure research at the source.
This is already reality at scale. Real-world federated implementations now handle datasets from hundreds of millions of patients across multiple continents. Networks like PCORnet ae link dozens of nodes, and researchers now cut cohort queries from weeks to seconds.
Lifebit was founded to make this model practical and repeatable. Our next-generation federated AI platform gives global institutions a production-ready way to collaborate on sensitive genomic and biomedical data securely, without compromising privacy, sovereignty, or performance. Components such as our Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence & Analytics Layer) together deliver the secure architecture needed for real-time, federated analysis.
What is a Federated Research Environment and How Does It Work?
A federated research environment (FRE) marks a paradigm shift from traditional research. Instead of centralizing huge datasets or manually sharing data copies—processes filled with security risks, delays, and spiraling cloud costs—an FRE works differently.
An FRE operates on the principle of “bringing computation to the data.” Sensitive, patient-level data never leaves its secure source. The typical workflow unfolds as follows:
- Discovery: A researcher uses a central portal to search for relevant datasets across the network. This search queries a centralized metadata catalog, which contains high-level, non-sensitive information about the data at each participating institution (or “node”).
- Cohort Definition: Based on the metadata, the researcher defines a cohort of interest (e.g., “females over 50 with a specific genetic marker and a diagnosis of type 2 diabetes”).
- Access Request: The researcher submits a formal proposal and an analysis plan, which is reviewed by a Data Access Committee (DAC) or equivalent governance body for each relevant node.
- Code Submission: Once approved, the researcher submits their analysis code (e.g., a Python script or R program) to the federated platform.
- Distributed Computation: The platform securely distributes the analysis code to each node holding the approved data. The code is executed locally within each institution’s secure environment (e.g., a Trusted Research Environment).
- Result Aggregation: Only aggregated, non-identifiable results (e.g., a statistical model, a p-value, or a patient count) are generated at each node. These partial results are then securely returned to a central point for aggregation into a final, combined result.
- Output Review: Before the final aggregated result is released to the researcher, it undergoes an automated or manual review (an “airlock” process) to ensure it does not pose a re-identification risk.
This approach avoids the vulnerabilities and high costs of moving and duplicating data in traditional models. In Lifebit’s federated model, approved users access data via linking technologies like Application Programming Interfaces (APIs) that connect directly to components such as our Trusted Research Environment (TRE) and Trusted Data Lakehouse (TDL).
To facilitate discovery across these distributed datasets, a centralized metadata layer provides information about the data at each node (for example, data types, patient counts, and high-level cohort characteristics) without revealing personally identifiable information. Researchers can query this metadata to find relevant cohorts and then submit analysis requests to the appropriate nodes.
| Feature | Traditional Centralized Data Warehouse | Federated Research Environment |
|---|---|---|
| Data Location | All data moved to a single repository | Data remains at source institutions |
| Security | Single point of failure, data in transit risks | Data never leaves source, reduced breach surface |
| Scalability | Limited by central infrastructure, costly to expand | Highly scalable, leverages distributed resources |
| Cost | High data transfer & storage, central infrastructure | Reduced data movement costs, optimized compute |
| Autonomy | Data custodians lose direct control over data | Data custodians retain full control and sovereignty |
| Data Sharing | Physical transfer, duplication | Virtual access, code-to-data approach |
| Privacy Risk | Higher re-identification risk due to centralization | Lower re-identification risk, privacy-by-design |
The Core Principles Guiding Federated Research
Federated research is more than a technical fix; it’s a collaborative ecosystem built on four principles to foster trust and accelerate findy: Adopt Standards, Build Trust, Enable Federation, and Accelerate Research.
-
Adopt Standards: Interoperability is key. Open standards like those from the Global Alliance for Genomics & Health (GA4GH) are crucial. They create consistent processes, reduce friction, and allow tools to connect to a broader ecosystem. This ensures datasets are accessible through the same APIs, streamlining research.
-
Build Trust: Trust is paramount. Best practices like the Five Safes Framework and structured engagement with leading organizations and the public help ensure transparency and ethical data use. This builds the “social license” to operate. As highlighted in research on Federalist principles for healthcare data networks, distributed governance and local control are essential for building trust.
-
Enable Federation: Federation is an ecosystem of independent organizations. It requires not just technology but also trust, autonomy, and governance. Lifebit’s platform supports this by enabling data administrators to set up collaborative workspaces in minutes and to define granular policies that respect local governance while still enabling cross-network analysis.
-
Accelerate Research: The ultimate goal is faster scientific findy. Federation achieves this by ensuring data safety, promoting inclusive research through wider data access, and boosting efficiency. Lifebit clients use our federated AI platform to cut time-to-insight, operationalize real-world evidence, and scale compliant research across regions.
How Federation Addresses Critical Privacy and Security Concerns
A key advantage of a federated research environment is how it inherently addresses privacy and security for sensitive data like genomic and healthcare information.
The “data stays in place” principle is a powerful privacy tool. Minimizing data movement drastically reduces the risk of breaches during transit. In Lifebit’s deployments, 100% of client data stays within the client’s controlled environment, whether that is an on-premises Secure Research Environment (SRE), a national TRE, or a cloud-hosted Trusted Data Lakehouse, helping maintain security compliance and reducing risk.
To further safeguard privacy, patient information is typically pseudonymized, meaning direct identifiers are replaced with artificial ones, making it incredibly difficult to link data back to an individual.
Beyond pseudonymization, advanced federated systems incorporate Privacy-Enhancing Technologies (PETs) for mathematical guarantees of privacy. Differential privacy, for instance, adds calibrated statistical noise to aggregated results, making it impossible to infer information about any single individual while preserving overall accuracy. Other techniques, like secure multi-party computation (SMPC), use cryptography to allow multiple parties to jointly compute a function over their private data without revealing it to each other. While computationally intensive, these methods offer the strongest privacy protections for certain analyses.
Strict controls like “airlock” systems are also in place. These gatekeepers review and approve all analysis results before release, ensuring only non-identifiable, aggregated data leaves the secure environment. This prevents inadvertent re-identification risks.
Data residency compliance is another critical aspect. With operations across London, New York, the United Kingdom, USA, Israel, Singapore, Canada, and Europe, Lifebit works within varied and stringent regulations. Our federated approach ensures data remains within its jurisdictional boundaries, supporting adherence to global standards like GDPR and HIPAA. This distributed control is fundamental to building public trust.
The Architecture of a Federated Research Environment
A federated research environment is a sociotechnical ecosystem of people, processes, and technology in a distributed network. It is not a single system but a set of interconnected components for secure, collaborative data analysis.
The architecture’s base consists of multiple “nodes,” each representing a data-holding institution like a hospital, biobank, regulator, or research center. These nodes keep their data in their own Secure Research Environments (SREs) or Trusted Research Environments (TREs), which can be deployed on-premises or in the cloud.
A coordinating center or central platform provides the infrastructure for discovery and orchestration. This central component doesn’t hold raw data but serves as the network’s switchboard. Its key functions include managing a unified metadata catalog for data discovery, handling researcher authentication and authorization, tracking access requests and approvals, and routing analytical queries to the appropriate nodes. It also aggregates the partial results returned from each node and manages the final output review process.
Interoperability is the linchpin that holds the network together. Common standards for data models, APIs, and communication protocols are essential for data to be analyzable across disparate nodes. The Global Alliance for Genomics & Health (GA4GH) provides a suite of crucial standards, including:
- Beacon API: Allows researchers to query whether a dataset contains a specific genetic variant without revealing further information.
- Data Repository Service (DRS): Provides a standardized way to identify and access data objects across different cloud and on-premise environments.
- Workflow Execution Service (WES): Defines a standard API for submitting and running analysis workflows, ensuring that the same analysis can be executed consistently at each node.
These standards allow analytical code to execute consistently across different institutions, even if their underlying storage systems vary.
Lifebit’s architecture extends this pattern with components such as the Trusted Data Lakehouse (TDL) and R.E.A.L. (Real-time Evidence & Analytics Layer), which sit alongside TREs to enable harmonized storage, federated AI/ML analytics, and real-time evidence generation across hybrid data ecosystems.
The Role of Secure Research Environments (SREs) as Trusted Nodes
Secure Research Environments (SREs)—also known as Trusted Research Environments (TREs) or Data Safe Havens—are the bedrock of a federated ecosystem. They are highly secure digital environments providing authorized researchers with remote access to analyze sensitive health data without ever releasing it.
These environments are built on the principle of “in-situ analysis,” where analysis happens where the data resides. This removes the need to physically share data, supporting the highest level of data governance. Data remains protected, and only authorized individuals can access and analyze it using a curated set of tools within the SRE.
SREs are often guided by the Five Safes framework to ensure:
- Safe People: Only authorized and trained researchers can access the data.
- Safe Projects: Data is used only for approved, societally beneficial research.
- Safe Settings: Access occurs within a secure, controlled computing environment.
- Safe Data: Data is de-identified or pseudonymized to protect privacy.
- Safe Outputs: All results are reviewed to prevent re-identification before release.
National-scale SREs and TREs can now support cohorts of hundreds of thousands of participants and beyond. Lifebit’s platform is proven to handle datasets from over 250 million patients across five continents, demonstrating the robustness required for national and international research.
Why Governance and Standards are the Foundation of Trust
In a distributed federated environment, robust governance and universal standards are foundational. Without them, the ecosystem cannot function securely, ethically, or efficiently.
Data use agreements are the legal and ethical contracts formalizing collaboration. They define who can access what data, for what purpose, and under what conditions, ensuring each data custodian maintains control.
Common Data Models (CDMs) like OMOP and HL7’s FHIR are technical specifications that standardize data structure. This harmonization is critical, allowing analytical code to be written once and run across multiple datasets, regardless of their original format. However, converting source data into a CDM via an Extract, Transform, Load (ETL) process is a significant undertaking. It requires deep domain expertise to map local coding systems to the CDM’s standardized vocabularies. This resource-intensive process must navigate data quality issues and institutional variations, but it is a crucial investment for achieving true cross-network interoperability.
The Global Alliance for Genomics & Health plays a pivotal role in developing these open standards, harmonizing the experience of finding and analyzing many datasets at once in compliance with international standards.
Data stewardship involves clear roles and responsibilities at each node for data quality, maintenance, and adherence to governance policies. This includes ensuring the CDM-mapped data is regularly updated and validated.
Finally, query review processes are integrated to ensure research questions align with approved protocols and that outputs pose no re-identification risk. This multi-layered approach to governance is what builds the trust necessary for successful federated research, and it is embedded in Lifebit’s federated governance model across TREs, TDLs, and real-time analytics layers.
From Months to Minutes: The Transformative Benefits of Federation
The shift to a federated research environment is not just about security; it is about fundamentally changing the pace, quality, and cost of scientific findy. Lifebit has seen how federation can accelerate research from a months-long endeavor to one measured in minutes.
Increased research velocity. Lifebit’s platform is designed to be 5x faster for identifying data, creating cohorts, and running analyses compared with traditional, manual data access workflows. This speed leads directly to faster insights, drug findy, and public health responses.
Improved data quality. By working with original, rich datasets rather than anonymized copies, researchers can conduct more granular and robust analyses. This direct access, combined with standardized data models, ensures more accurate and reliable insights.
Seamless collaboration. Researchers can collaborate on complex projects without data transfer logistics. Our platform lets administrators create collaborative workspaces in under 10 minutes, connecting TREs, TDLs, and analytical tools, and fostering an efficient environment.
Unparalleled scalability. Federation enables population-level studies by virtually unifying vast datasets. Lifebit’s federated AI platform handles data from over 250 million patients across 5 continents, enabling studies previously impossible due to data access limits.

Slashing Costs and Boosting Research Efficiency
Federated research environments also deliver significant economic advantages by slashing costs and boosting efficiency.
One of the most immediate benefits is eliminating data transfer costs. Moving large datasets is expensive due to egress fees and bandwidth needs. As an AWS blog post on the movement of large datasets is often very costly for researchers. notes, keeping data at its source avoids these prohibitive costs.
Cloud optimization is another key area. Lifebit’s platform, often hosted on providers like AWS, offers a flexible, pay-as-you-go model. We automate recommendations for the appropriate AWS instance for batch workflows to ensure efficient resource allocation. The AWS Instance Cost Calculator is a valuable tool for estimating costs. If researchers are unsure about analysis time, cost, or instance selection, they can book an appointment with a specialist for guidance.
Efficiency gains extend to the research workflow itself. Lifebit allows users to build precise cohorts in as little as 30 seconds, drastically reducing time spent on data wrangling and freeing up researchers to focus on analysis.
Powering More Inclusive and Higher-Quality Science
Federated research also fundamentally improves the inclusivity and quality of science.
By enabling secure access to distributed datasets, federated environments provide access to diverse, representative datasets. This is crucial in healthcare, where outcomes vary by demographic and geography. Unlike centralized datasets that often suffer from selection bias, federation allows researchers to access data from multiple regions (including London, New York, the United Kingdom, USA, Israel, Singapore, Canada, and Europe). This reduces research bias and ensures medical advancements are equitable.
Furthermore, federated environments excel at combining multi-modal data. Clinical, imaging, and genomic data can be integrated and analyzed together without leaving their secure source. National TREs and large research programs, with access to clinical, imaging, and genomic information from hundreds of thousands of participants, demonstrate this power in practice. This holistic view enables deeper insights into disease, treatment efficacy, and personalized medicine.
The ability to analyze real-world data at scale contributes significantly to real-world evidence. This evidence, derived from routine clinical practice, complements traditional trials by providing insights into how treatments perform in diverse populations, ultimately improving patient care and enabling proactive pharmacovigilance.
Navigating the Problems: Challenges and Practical Considerations
While compelling, implementing federated research environments is complex. Navigating this frontier means addressing technical, organizational, and regulatory challenges.
Technical complexity arises from integrating disparate systems, ensuring interoperability, and managing distributed compute. Legacy systems and varied data formats at each node create real-world problems. For instance, one institution might use a different identity provider (e.g., Azure AD vs. Okta), requiring complex authentication federation. Network firewalls and security policies at each hospital or research center can block the communication required for federated queries, necessitating careful negotiation and configuration with local IT teams. While containerization technologies like Docker help package analysis tools, ensuring they run consistently across heterogeneous compute environments (on-premise clusters, different cloud providers) remains a significant engineering challenge.
Organizational alignment is crucial. Bringing together independent institutions, each with its own governance and priorities, requires significant coordination to align goals and ensure consistent participation.
Regulatory variation across jurisdictions (like the USA, Canada, Europe, and other regions where Lifebit operates) adds another layer of complexity. For example, Europe’s GDPR has strict rules on the transfer of personal data outside the EU, while the USA’s HIPAA governs protected health information but has different consent and de-identification standards. A federated network spanning these regions must have a flexible governance framework that can enforce the strictest applicable rule for any given query, a concept known as policy synthesis. Navigating differing patient consent models and data-sharing permissions requires careful legal and ethical navigation to ensure compliance across the network.
Lifebit’s approach is to provide a federated framework that can be configured to local policies while still enabling cross-border analysis where legally permitted, helping organizations move faster without compromising compliance.
The ‘Invisible Work’ of Implementing a Federated Research Environment
A vastly underestimated aspect of establishing a federated research environment is the “invisible work” of infrastructuring. This goes beyond technology deployment to the extensive relational and organizational effort needed to make a distributed network function.
A case study of PCORnet®️, a national federated network in the USA, revealed that most of the work (57%) was relational, not technical (43%). This “invisible work” included aligning governance, motivating contributions, coordinating resources, and supporting researchers. The Research data query-response workflows for PCORnet®️ illustrate this multi-stakeholder process.
To prepare for node development, organizations should consider these pragmatic questions:
- How will local teams access and review network functional requirements and technical specifications?
- How will existing expertise and local data governance be aligned with network technical requirements?
- Are resources available to engage contractors if internal technical capacity is limited?
- How will conflicts between local policies and network requirements be addressed?
- How does the organization commit to embedding infrastructural components into its own infrastructure?
- How will the project team motivate the contribution of internal resources beyond the start-up budget?
- How will decision-makers who need to approve actions be engaged?
- What processes are required to support ongoing operations of the local system?
- What data stewardship processes will be used to review query results?
- How will patients, caregivers, and other key stakeholders be engaged for input?
Lifebit works with governments, public health agencies, and biopharma to address this invisible work through implementation playbooks, governance templates, and long-term partnership models that help sustain federated networks beyond initial funding cycles.
Practical Costs and Resource Planning
While federation offers long-term efficiencies, it involves upfront costs and requires careful resource planning.
Initial setup fees: Costs may include platform licensing, secure environment configuration, and IT integration, including API and connector development.
Ongoing compute costs: As a primary expense, analyses run on cloud platforms like AWS incur pay-as-you-go costs for compute, storage, and networking. Large analyses can be costly. For example, national-scale Trusted Research Environments may charge for initial AWS compute usage.
Need for specialized expertise: Building and maintaining a federated environment requires a multidisciplinary team. Key roles often include:
- Data Engineers: For the ETL process and data pipeline management.
- Bioinformaticians/Data Scientists: To design analyses and support researchers.
- Cloud/Infrastructure Engineers: To manage the secure TRE infrastructure and connectivity.
- Governance and Compliance Specialists: To handle data use agreements, access committees, and regulatory compliance.
- Project/Network Managers: To coordinate between partners and manage stakeholder alignment.
Data harmonization effort: Converting disparate datasets into a common data model represents a significant upfront investment of time and expert resources, but it pays dividends in long-term efficiency.
Training and education: Researchers, custodians, and administrators need education on federated principles, platform use, and governance protocols. Lifebit supports this with onboarding, documentation, and ongoing enablement. For detailed cost estimations, we encourage researchers to book an appointment with a Lifebit Solutions Specialist.
The Future is Federated: Embracing the Next Paradigm of Research
The federated frontier is just beginning, but its potential is clear. By breaking down data silos while upholding strict privacy standards, federated research environments accelerate findy, cut costs, and empower a new era of inclusive, high-quality science.
Federation dramatically increases research velocitymaking analysis 5x faster and cutting cohort building from weeks to seconds. It eliminates data transfer costs, optimizes cloud resources, and enables access to diverse, population-scale datasets. This reduces bias and generates richer real-world evidence.
Looking ahead, the future of research is federated. We see an evolution towards Learning Health Systems, where research insights improve clinical care, and clinical data informs research in a virtuous cycle that drives better patient outcomes.
The rise of empowering AI agents within federated environments will further revolutionize findy. These agents can operate autonomously across distributed data, integrating diverse information to accelerate scientific breakthroughs, power real-time safety surveillance, and support regulatory-grade evidence generation.
Regulatory initiatives like the European Health Data Space (EHDS) show the global momentum for secure, cross-border health data sharing. Lifebit’s EHDS-ready Trusted Research Environment solution is designed to support compliance with emerging EHDS regulations, paving the way for international collaboration.
At Lifebit, our federated platform is at the forefront of this revolution. We provide a next-generation federated AI platform for secure, real-time access to global biomedical and multi-omic data. With built-in harmonization, advanced AI/ML, and federated governance, we power large-scale research and pharmacovigilance for biopharma and governments in the UK, USA, Canada, Europe, Israel, Singapore, and beyond. Components such as our TRE, TDL, and R.E.A.L. layer work together to deliver real-time insights, AI-driven safety surveillance, and secure collaboration across hybrid data ecosystems. We offer a 100% client results guarantee, ensuring our partners achieve their research objectives.
The era of data-intensive science is here, and federation is its guiding principle. The organizations that act now will set the standards for how biomedical data is used safely and effectively over the next decade.
Discover how to implement a federated biomedical data platform

