Why Your Clinical Data is Out of Tune and How to Fix It

Why Clinical Data Sits in Silos—And What It Costs You
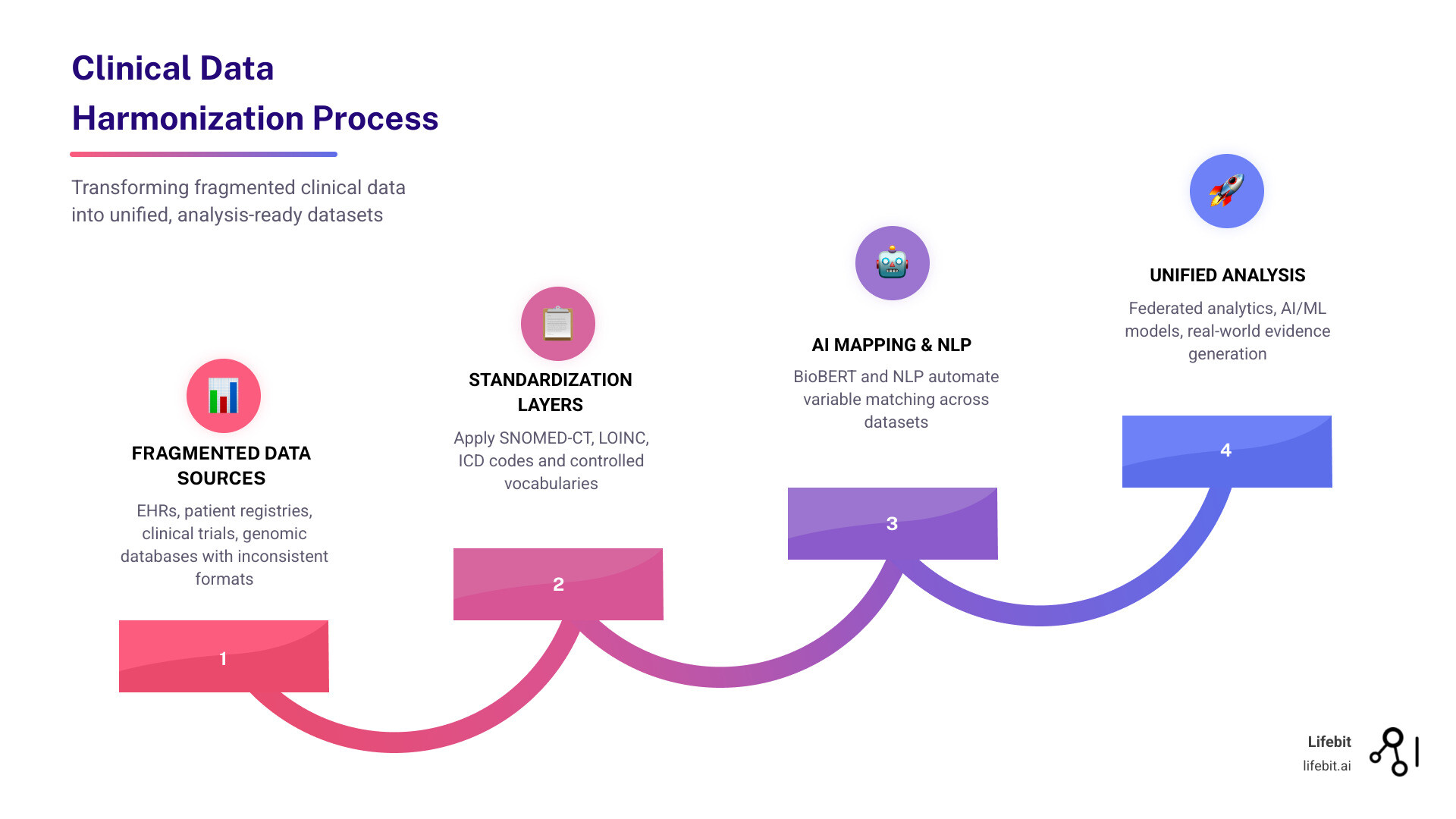
Clinical data harmonization is the process of integrating disparate health data from multiple sources—like electronic health records (EHRs), patient registries, clinical trials, and genomic databases—into a unified, standardized format that enables cross-study analysis and real-world evidence generation.
What you need to know:
- The problem: Clinical datasets use inconsistent variable names, coding systems, and formats—making cross-institutional research nearly impossible without months of manual reconciliation
- The impact: A single Phase III trial generates 3.6 million data points, but 30% of data is often excluded because it can’t be harmonized
- The solution: Modern approaches combine standardized vocabularies (SNOMED-CT, LOINC, ICD codes) with AI-driven methods like NLP and BioBERT to automate mapping
- The payoff: Harmonized data enables federated analytics, accelerates cohort recruitment by 60%, and unlocks AI/ML applications for predictive modeling
Modern healthcare sits on a goldmine of clinical data.
But here’s the problem: that data doesn’t speak the same language.
One hospital calls it “SystolicBP.” Another uses “SBPvisit1.” A third logs it as “Blood Pressure – Systolic Reading.” Same measurement. Three different names. Multiply that across thousands of variables, dozens of institutions, and multiple countries—and you’ve got a data harmonization nightmare that’s costing pharma companies months of delay and researchers millions in wasted effort.
This isn’t just an inconvenience. When the FDA’s SHIELD program analyzed laboratory data integrity across the healthcare ecosystem, they found that data maintains only 22-68% accuracy after a single round-trip through the data lifecycle. The NHLBI’s CONNECTS network discovered that retrospective harmonization delays data sharing by 2-7 months per study. And when diabetes status descriptions vary across cohort studies, cross-cohort risk prediction becomes impossible.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, where we’ve spent over 15 years solving clinical data harmonization challenges across genomics, biomedical data platforms, and federated research environments for public sector institutions and pharmaceutical organizations worldwide. Throughout my career—from building Nextflow, the breakthrough workflow framework used globally in genomic data analysis, to leading research at the Centre for Genomic Regulation—I’ve seen how poor clinical data harmonization blocks precision medicine, stalls drug discovery, and prevents AI models from reaching their full potential.
Key clinical data harmonization vocabulary:
Clinical Data Harmonization: Stop Losing 30% of Trial Data to Messy Formats
clinical data harmonization definition; research efficiency; data interoperability; [Scientific research article]; [IMAGE]
At its core, clinical data harmonization is about creating a “universal translator” for medical information. It is the technical and semantic work required to ensure that data collected in different settings-using different software, different languages, and different medical terminologies-can be pooled together for a single analysis.
Why is this essential? Because modern healthcare research is increasingly “big data” research. A single Phase III clinical trial now produces an average of 3.6 million data points. However, the real power lies in looking across trials, across hospitals, and across borders.
Without harmonization, you are looking at a fragmented mosaic. You might have access to 187 million patient records globally, but if 90% of those are “ex-US” and use different coding schemes than your domestic data, you cannot build a robust predictive model. Harmonization transforms these silos into a “Trusted Data Factory,” where analysis-ready data products are reusable and scalable.
This process is the bedrock of the FAIR guiding principles-making data Findable, Accessible, Interoperable, and Reusable. Without it, the “interoperable” and “reusable” parts simply do not happen.
The Primary Challenges of Harmonizing Clinical Data from Diverse Sources
EHR heterogeneity; registry silos; metadata variability; [Scientific research article]; [INFOGRAPHIC]
If harmonization were easy, we’d have solved it decades ago. The reality is that clinical data is messy, idiosyncratic, and deeply rooted in the specific context where it was collected.
Tackling Inconsistent Variable Names and Textual Descriptions
variable naming conventions; cardiovascular cohorts; FHS; MESA; ARIC; [Scientific research article]
The most common barrier is the sheer variety of naming conventions. Researchers often use custom shorthand that makes sense within a single study but is opaque to outsiders.
Consider a study of cardiovascular health. Researchers analyzed 885 variable descriptions from three major cardiovascular cohort studies: the Framingham Heart Study (FHS), the Multi-Ethnic Study of Atherosclerosis (MESA), and the Atherosclerosis Risk in Communities (ARIC) study. They found these 885 variables actually represented 64 core concepts across 7 groups.
Manually mapping “diabetesstatusv1″ to a standardized concept like “Diabetes Mellitus Diagnosis” across thousands of columns is a Herculean task. Traditional manual harmonization is time-consuming, error-prone, and fundamentally unscalable for the petabyte-scale datasets many organizations handle today.
The Unit Harmonization and Missing Data Hurdle
lab value units; missingness; domain-specific variations; [Scientific research article]
Even when you agree on the variable name, the values inside the column might not match. One lab might report glucose in mg/dL, while another uses mmol/L. Without precise unit harmonization, an AI model can draw the wrong conclusion simply because of a unit conversion error.
Furthermore, different studies have different patterns of “missingness.” Some data is missing at random; some is missing because a specific test was not relevant to that cohort. Handling these domain-specific variations requires a hybrid approach that looks at both the metadata (the descriptions of the data) and the patient-level data itself to ensure robustness.

From Manual Consensus to AI: The Evolution of Harmonization Approaches
;manual vs automated; AHRQ OMF; machine learning; [Scientific research article]; [TABLE]
Historically, harmonization was a social process, not just a technical one. It involved rooms full of experts debating definitions until they reached a consensus.
Traditional Approaches: AHRQ’s Outcomes Measures Framework (OMF)
;consensus building; standardized outcome libraries; [AHRQ information page]
The Agency for Healthcare Research and Quality (AHRQ) led the way with the Outcomes Measures Framework (OMF). This project brought together registry holders, EHR developers, and clinicians to develop standardized outcome measure libraries for five key clinical areas:
- Atrial fibrillation
- Asthma
- Depression
- Non-small cell lung cancer
- Lumbar spondylolisthesis
By building these consensus definitions, they created a foundation for registries to talk to each other. You can view these public outcome libraries here. While effective, this manual “working group” model can take years to produce standards for just a handful of conditions.
Changing Scalability with NLP and BioBERT Clinical Data Harmonization
;NLP; BioBERT embeddings; FCN model; contrastive learning; [Scientific research article]
The game-changer is the shift toward automation. Recent studies have demonstrated that Natural Language Processing (NLP) and machine learning models can do the heavy lifting of mapping free-text descriptions to standardized concepts.
Using domain-specific models like BioBERT, researchers have achieved incredible performance metrics. In one study, a Fully Connected Network (FCN) model trained on 58,890 generated sentence pairs achieved:
- AUC: 0.99 (95% CI 0.98-0.99)
- Top-1 Accuracy: 80.51%
- Top-5 Accuracy: 98.95%
This indicates that AI can identify whether two variables from different studies mean the same thing with near-perfect accuracy. This level of ai for data harmonization is what allows us to scale from harmonizing hundreds of variables to millions.
Standardized Vocabularies and Common Data Models: The Shared Language of Healthcare
;SNOMED-CT; LOINC; ICD; OMOP; PCORnet; [Scientific research article]; [IMAGE]
To harmonize data, you need a target destination—a “Common Data Model” (CDM) and a “Controlled Vocabulary.”
Mapping to SNOMED-CT, LOINC, and ICD Codes
;controlled terminologies; RxNorm; NDC; [Scientific research article]
Standardized vocabularies act as the dictionary for harmonization:
- SNOMED-CT: Used for clinical findings, symptoms, and diagnoses.
- LOINC: The gold standard for identifying laboratory tests and observations.
- ICD-10/11: Used for billing and epidemiological tracking of diseases.
- RxNorm/NDC: Standard codes for clinical drugs.
The FDA’s SHIELD initiative emphasizes that using these codes isn’t enough—they must be applied systematically. For example, the enzyme Aspartate transferase (AST) can be under-measured if the test method isn’t supplemented with Vitamin B6. Harmonization requires knowing which LOINC code was used to ensure the results are truly comparable.
The Role of OMOP, PCORnet, and Sentinel CDMs
;Common Data Models; OHDSI; i2b2; ACT; [Scientific research article]
Once the individual terms are coded, the data needs to be structured into a Common Data Model. These models define how tables (like “Person,” “Procedure,” or “Drug Exposure”) relate to each other.
- OMOP (Observational Medical Outcomes Partnership): Used by the OHDSI network at over 90 sites worldwide. It is the most popular model for observational research.
- PCORnet: Supported by over 80 institutions, focusing on patient-centered outcomes.
- i2b2/ACT: Used at over 200 sites, often for cohort discovery and finding patients for clinical trials.
Using a CDM allows a researcher to write one query and run it across dozens of different hospital databases simultaneously. This is the foundation of from chaos to clarity: implementing OMOP.
The Business Case for Harmonization in Pharma R&D and Clinical Trials
;Pharma R&D cohort recruitment; real-world evidence; [Deloitte blog post]
For pharmaceutical companies, clinical data harmonization isn’t just a technical goal—it’s a massive competitive advantage.
Accelerating Cohort Recruitment and Cross-Cohort Analyses
;recruitment efficiency; NSCLC; rare diseases; [Scientific research article]
One of the biggest bottlenecks in drug development is finding the right patients. By integrating phenotype, omics, and clinical data in a federated environment, organizations have seen cohort recruitment accelerated by 60%.
In rare disease research, where a single institution might only have two or three relevant patients, harmonizing data across 12 sites or more is the only way to reach a statistically significant sample size. Standardizing longitudinal data from 1,000 patients across different countries allows for the kind of “mega-analysis” that regulators and payers now demand.
Powering AI/ML Applications and Real-World Evidence (RWE)
;predictive modeling; 21st Century Cures Act; RWD; [Scientific research article]
The 21st Century Cures Act encourages the use of Real-World Data (RWD) to augment traditional randomized controlled trials (RCTs). But AI is only as good as the data it’s fed.
Harmonized data enables:
- Predictive Modeling: Using laboratory value trajectories to predict severe outcomes, as seen in COVID-19 studies across 342 hospitals.
- Safety Surveillance: Real-time monitoring of adverse events across global populations.
- Precision Medicine: Linking clinical outcomes to genomic data to identify which patients will respond best to a specific molecular agent.
Without a solid foundation of health data harmonization, your AI models will be “hallucinating” on inconsistent data inputs.
Best Practices and Future Directions for Sustainable Data Governance
;few-shot learning; LLMs; multi-omics; Lifebit; [Scientific research article]
As we look toward 2026 and beyond, the field is moving away from retrospective “clean-up” and toward “harmonization by design.”
Advancing Clinical Data Harmonization with Few-Shot Learning
;Few-shot learning; LLMs; scalability; [Scientific research article]
The next frontier involves Large Language Models (LLMs) and few-shot learning. While BioBERT requires thousands of labeled examples, few-shot learning allows models to learn how to harmonize new clinical domains with only a handful of examples. This will be critical for emerging fields like cell and gene therapy, where the data types are entirely new and historical labels don’t exist.
Multi-Omics Integration and the Role of Lifebit
;Lifebit; federated AI; TRE; TDL; R.E.A.L.; multi-omics
At Lifebit, we believe the future of clinical data harmonization is federated. You shouldn’t have to move sensitive patient data to harmonize it. Our platform allows you to bring the “harmonization engine” to the data, wherever it sits—in London, New York, Singapore, or beyond.
Our Trusted Research Environment (TRE) and Trusted Data Lakehouse (TDL) provide the infrastructure to:
- Ingest fragmented, multi-modal data (imaging, EMR, genomics).
- Harmonize it automatically using AI-driven mastering and standard CDMs like OMOP.
- Analyze it in real-time through our R.E.A.L. (Real-time Evidence & Analytics Layer).
This approach ensures that your data is not just “integrated” but truly “analysis-ready,” maintaining the highest standards of security and data-harmonization-overcoming-challenges.
Sustainable harmonization requires more than just code; it requires governance. This means involving stakeholders—clinicians, patients, and IT directors—early in the process and building “data libraries” that preserve the provenance of every change.
Is your data ready for the AI era?
If your research is still being held back by manual data cleaning and incompatible formats, it’s time to tune your clinical data strategy. By adopting automated harmonization and federated governance, you can stop fighting with your data and start finding the insights that save lives.
To see how we can help you build your own Trusted Data Factory, discover the Lifebit platform.

