From Data to Discovery: A Guide to AI in Biopharma Analytics

How Lifebit Cuts Target Discovery Timelines 90% with AI-Powered Omics Analytics
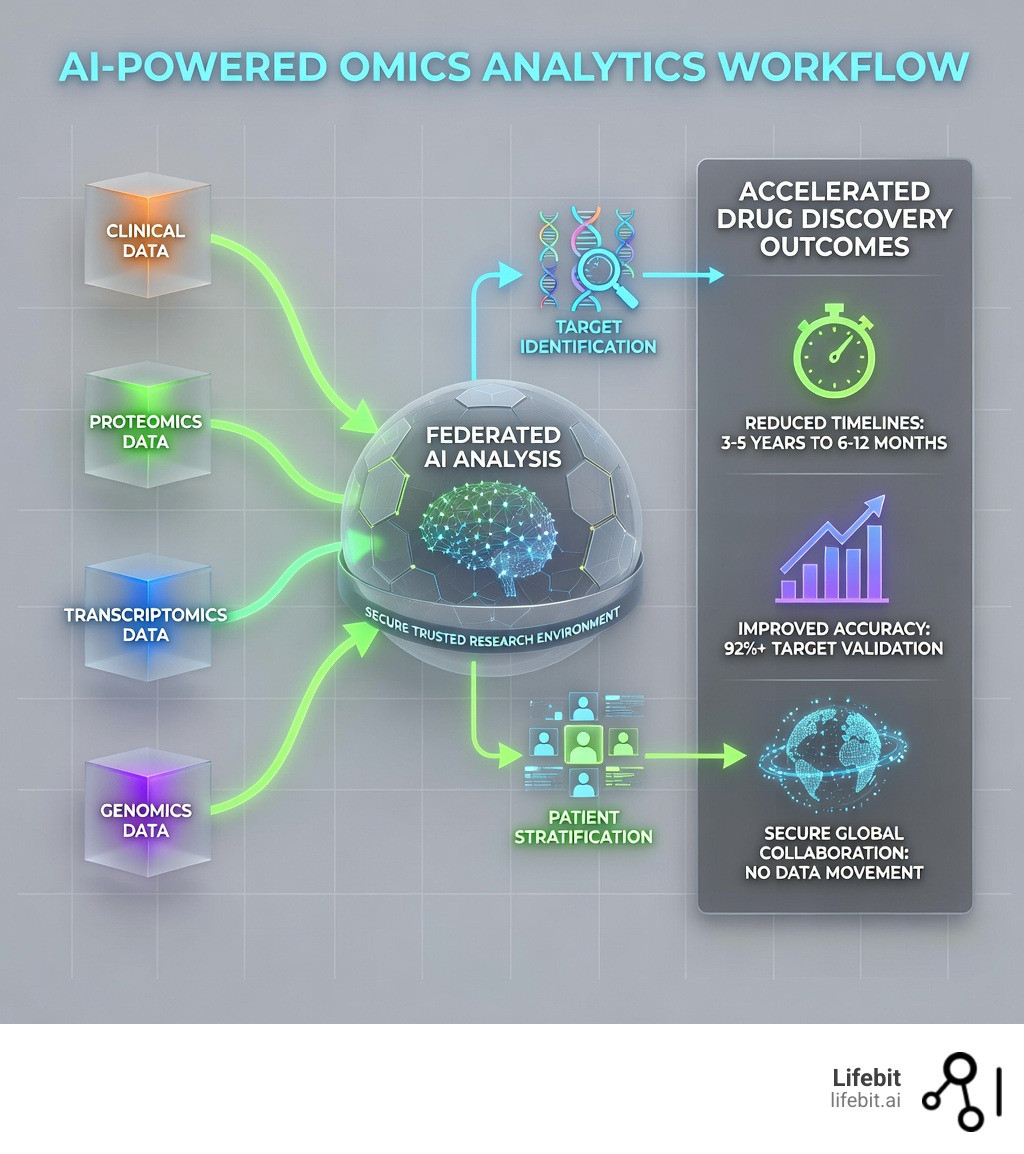
AI-powered omics analytics is revolutionizing how researchers connect genetic information to disease mechanisms and drug targets. For decades, the biological sciences operated under a hypothesis-driven model, where researchers would spend years investigating a single gene or protein. Today, the paradigm has shifted to a data-driven approach. We are no longer limited by our ability to generate data, but by our ability to interpret it. This is where artificial intelligence becomes the indispensable engine of modern drug discovery.
What It Is:
- Integration of Multi-Layered Data: AI-powered omics analytics involves the simultaneous analysis of genomics (DNA), transcriptomics (RNA), proteomics (proteins), and metabolomics (metabolites). By layering these datasets, AI can identify correlations that are invisible to the human eye or traditional statistical methods.
- High-Throughput Platforms: These systems analyze millions of samples across diverse populations to identify novel drug targets and biomarkers with high statistical power.
- Federated Architectures: Modern systems utilize federated learning, enabling secure analysis across global datasets (like UK Biobank or Genomics England) without moving sensitive patient data from its original secure location.
Key Capabilities:
- Target Identification & Prioritization: AI models can rank thousands of potential genes based on novelty, safety, and “druggability” across 1.3M+ disease-specific samples, filtering out candidates likely to fail in clinical trials.
- Multi-Modal Integration: Beyond molecular data, these platforms ingest clinical trial results, electronic health records (EHR), scientific publications, and real-world evidence (RWE) to create a 360-degree view of patient biology.
- Accelerated Discovery Pipelines: By using predictive modeling, researchers can reduce the target validation phase from an average of 3-5 years down to just a few weeks or months.
Why It Matters:
- Overcoming Eroom’s Law: Despite technological advances, the cost of developing new drugs has historically doubled every nine years (Eroom’s Law). AI-powered omics analytics is the first technology with the potential to reverse this trend by increasing the probability of success (PoS) at every stage.
- Precision Medicine: Traditional “one-size-fits-all” medicine is being replaced by therapies tailored to a patient’s specific molecular profile. AI models now achieve 90%+ accuracy in predicting how specific patient subgroups will respond to a drug.
- Global Health Security: Federated networks have processed 1.6M+ viral genomes for pandemic surveillance, allowing for real-time tracking of mutations without compromising national data privacy laws.
The challenge for pharma, public health agencies, and research institutions isn’t whether to adopt AI-powered omics—it’s how to implement it securely, at scale, and with clear ROI. Most organizations struggle with three critical bottlenecks: siloed data that can’t be harmonized across different cloud providers, expensive multi-omics datasets that don’t answer strategic questions, and “black box” AI models that researchers can’t trust or interpret for regulatory filings.
This is where strategy-first implementation becomes essential. Organizations that jump straight to generating massive omics datasets often waste millions on data that never informs go/no-go decisions. The ones succeeding right now are those who define their translational objectives first, ensure data quality from biospecimen collection through analysis, and adopt federated platforms that enable collaboration without moving sensitive patient data.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, where we’ve built a federated genomics platform that powers secure AI-powered omics analytics for pharmaceutical companies and public health institutions globally. Before founding Lifebit, I contributed to Nextflow—the workflow framework now used worldwide for genomic data analysis—and conducted research at the Centre for Genomic Regulation building tools for precision medicine.
Throughout this guide, we’ll show you exactly how leading biopharma teams are using AI to compress time-to-insight from months to minutes, how federated systems solve the data access problem without compromising compliance, and which specific tools and methods are delivering validated clinical impact right now.
Access 1.3M Samples Instantly via AI-Powered Omics Analytics

To truly master AI-powered omics analytics, a platform must do more than just “crunch numbers.” It needs to act as a bridge between raw data and clinical action. At Lifebit, we focus on integrating multi-omics data—including genomics, transcriptomics, and proteomics—into a unified Trusted Research Environment (TRE).
The Power of Massive Knowledge Bases
Our platform leverages vast knowledge bases, such as the PandaOmics system, which contains 1.3M disease-specific omics samples, 15K clinical-stage compounds, and over 47M publications. This isn’t just a library; it’s an active discovery engine. By using multi-modal AI models, we can prioritize genes based on novelty, safety, and druggability.
For researchers in academia, access to these high-level tools is becoming more affordable. Specialized pricing as low as $199 per month for platforms like PandaOmics ensures that even smaller labs can compete in the precision medicine race. This democratization of data is critical because breakthroughs often come from unexpected places—a small lab in one country might find the key to a rare disease that a major pharma company overlooked.
Unlocking Actionable Insights with Lifebit’s Secure, Scalable Platform
The “secret sauce” of modern analytics lies in the ability to run next-generation sequencing (NGS) analysis and RNA-Seq reanalysis without the headache of manual data cleaning. Our Trusted Data Lakehouse (TDL) provides normalized omics collections, allowing you to skip the tedious preprocessing and go straight to pathway analysis.
Consider the technical burden of traditional methods: a researcher would typically spend 80% of their time on data wrangling—cleaning CSV files, converting file formats (BAM to FASTQ), and ensuring metadata consistency. AI-powered omics analytics automates this entire pipeline. Whether you are identifying a small-molecule TNIK inhibitor for fibrosis or predicting dual-purpose targets for aging and disease, our platform provides the data-driven evidence needed to back up your hypotheses. Real-time insights mean you can ask questions in natural language and receive biologically grounded answers, moving from variant to target in record time.
Scaling AI-Powered Omics Analytics for Global Collaboration
Data is often stuck in silos—locked away in different hospitals, countries, or clouds. This is the “Data Gravity” problem: as datasets grow to petabyte scale, they become too heavy to move. We solve this through federated networks. Instead of moving sensitive patient data (which is a regulatory nightmare under GDPR and HIPAA), we move the AI models to the data.
This approach is already proven at the highest levels of public health. Through federated networks, over 1.6M SARS-CoV-2 genomes were processed for pandemic surveillance. By adhering to GA4GH open standards, we ensure that AI-powered omics analytics can scale across global collaborations while maintaining strict privacy and federated governance. This means a researcher in London can run an analysis on a dataset in New York without the data ever leaving its secure server, ensuring compliance while maximizing scientific output.
Hit 90% Accuracy in Drug Response with AI-Powered Omics Analytics
In the fight against refractory diseases like Triple-Negative Breast Cancer (TNBC), traditional drug discovery often feels like looking for a needle in a haystack—while the haystack is on fire. AI-powered omics analytics provides the fire extinguisher by identifying specific molecular vulnerabilities that were previously hidden.
The Rise of Pyroptosis Therapy
Recent research has highlighted the power of pyroptosis therapy—a form of programmed cell death that “ignites” the immune system. Unlike apoptosis, which is a quiet cell death, pyroptosis releases pro-inflammatory cytokines that alert the immune system to the presence of cancer. Using advanced AI models, researchers have identified 9 prognostic pyroptosis genes that can predict recurrence-free survival in TNBC patients. This allows clinicians to identify which patients will benefit from immunotherapy and which require a different approach.
Accelerating Discovery with the BFReg-NN Model
One of the most exciting breakthroughs is the BFReg-NN (Biological Factor Regulatory Neural Network) model. Unlike “black box” models that offer no explanation for their predictions, BFReg-NN integrates biological knowledge, such as protein-protein interaction (PPI) networks, to predict drug pair effects. It doesn’t just look for statistical patterns; it respects the laws of biology.
In a landmark study, this model achieved a c-index of 0.90 for the combination of Mitoxantrone (MIT) and Gambogic Acid (GA). The c-index, or concordance index, is a measure of predictive accuracy; a score of 0.90 is exceptionally high in the context of complex biological systems. When tested in the lab, 12 out of 12 drug pairs predicted by the AI successfully induced pyroptosis in TNBC cells. This level of accuracy allows us to optimize drug ratios—like the 1:1.5 MIT:GA mass ratio—before ever entering a wet lab, significantly reducing clinical trial costs and sparing patients from ineffective treatments.
Breakthroughs in Single-Cell and Spatial Omics
The future of oncology is single-cell. Bulk sequencing (analyzing a whole tumor at once) often masks the diversity of cells within that tumor. Tools like scEMAIL are advancing how we annotate cell types without needing source data, while DGMP helps us identify cancer driver genes even when they haven’t mutated, by looking at their regulatory influence.
Furthermore, our spatial omics pipeline integrates transcriptomic data with histopathological images (using tools like TIST). This allows us to see exactly where a drug is working within a tumor’s ecosystem. Is the drug reaching the core of the tumor, or is it being blocked by the stroma? This level of detail is critical for biomarker discovery and understanding the hallmarks of aging. By mapping the “neighborhood” of a cell, AI-powered omics analytics provides a roadmap for the next generation of precision therapies.
Stop Wasting Millions: High-ROI AI-Powered Omics Analytics Strategy
If you treat AI as a magic wand, you’re going to be disappointed. To maximize ROI, organizations must adopt a strategy-first approach. The biggest cost in omics isn’t the AI software—it’s the generation of high-quality biospecimens and datasets. If the input data is flawed, the AI’s output will be equally flawed, leading to expensive failures in Phase II and III clinical trials.
Phenotypic Drug Discovery and Compound Optimization
Over 90% of successful drugs started with phenotypic discovery—observing how a drug affects a whole cell or organism rather than just a single isolated protein. By using AI-powered omics analytics to perform similarity learning on transcriptional signatures, we can identify how a compound affects the entire cell. This is how we move from “maybe this works” to “we know why this works.”
For example, the development of MG@PM nanococrystals (MIT and GA coated in platelet membranes) showed 6.5x higher tumor accumulation than standard treatments. AI didn’t just find the drugs; it helped optimize the delivery mechanism. By simulating how these nanococrystals interact with the cell membrane, researchers could refine the coating to ensure maximum penetration and minimal side effects. This is the difference between a drug that works in a petri dish and one that works in a human body.
Validating Clinical Impact: Lifebit Case Studies
Our collaborations have shown that AI can cut query turnaround times from weeks to minutes. This speed isn’t just about convenience; it’s about the pace of scientific discovery.
- Longevity Science: We are currently working with partners to predict dual-purpose targets—genes that, when modulated, can treat specific diseases like Alzheimer’s while simultaneously slowing the underlying biological aging process. This “Geroscience” approach could extend the human healthspan significantly.
- Infectious Disease: During the height of the COVID-19 pandemic, our platform processed hundreds of thousands of viral sequences in real-time. This allowed public health officials to track the emergence of the Delta and Omicron variants weeks before they became dominant, enabling faster policy responses.
- Oncology: By unifying registry data for 60K+ patients, we’ve allowed researchers to self-serve natural language queries. Instead of waiting for a bioinformatics team to run a script, a clinician can ask, “Which patients with this specific mutation responded best to checkpoint inhibitors?” and get an answer in seconds.
The Economic Argument for AI-Powered Omics
The average cost to bring a drug to market is now estimated at $2.6 billion. A significant portion of this cost is due to the 90% failure rate in clinical trials. If AI-powered omics analytics can improve the success rate by even 10%, the savings to the healthcare system and the pharmaceutical industry would be in the hundreds of billions of dollars. This is why the ROI on these platforms is so high; they don’t just save time, they mitigate the most significant financial risk in the industry.
Kill the ‘Black Box’ Problem in AI-Powered Omics Analytics
The biggest hurdle to adopting AI-powered omics analytics is trust. If a researcher can’t see why an AI made a prediction, they won’t risk a multi-million dollar trial on it. Regulatory bodies like the FDA and EMA are also increasingly demanding “explainability” in AI models used for clinical decision-making.
Solving the ‘Black Box’ Problem in Machine Learning
We are moving toward explainable ML (XAI). Traditional deep learning models are often criticized for being opaque. However, new tools like SOPHIE use generative neural networks to separate common transcriptional responses (the “noise”) from specific, disease-related ones (the “signal”). This makes it easier for a biologist to see the true biological driver behind a prediction.
By using model interpretability tools, such as SHAP (SHapley Additive exPlanations) or LIME (Local Interpretable Model-agnostic Explanations), we can assign a “weight” to each gene’s contribution to a prediction. If an AI says a patient will respond to a drug, it can now point to the specific expression levels of five key genes that led to that conclusion. This allows researchers to optimize models for reproducibility, ensuring that the insights you get today are still valid tomorrow across different patient cohorts.
Prioritizing Data Quality and Seamless Integration
“Garbage in, garbage out” is the mantra of the omics world. Successful implementation requires more than just a good algorithm; it requires a robust data strategy:
- FAIR Data Principles: Data must be Findable, Accessible, Interoperable, and Reusable. Without these standards, AI models cannot effectively learn from historical data.
- Harmonization and Batch Effect Correction: When combining data from different labs or sequencing machines, “batch effects” can create false signals. Using Adversarial Autoencoders, we can “neutralize” these effects, ensuring the AI is looking at biological differences, not technical ones.
- Biospecimen Strategy: The quality of the original tissue sample is paramount. Ensuring that samples are collected, frozen, and processed consistently is the only way to support deep proteomics and genomic analysis. AI can even help here, by flagging samples that appear to be outliers or of low quality before they enter the analysis pipeline.
Slash Clinical Trial Costs: AI-Powered Omics Analytics FAQ
How does Lifebit’s AI improve target identification accuracy?
By integrating multi-modal data—molecular signals, PPI networks, and vast literature knowledge bases—our AI filters out the noise. Traditional methods often find correlations that don’t hold up in vivo. Models like BFReg-NN achieve up to 90% accuracy by respecting biological hierarchies and regulatory networks rather than just looking for simple statistical correlations. This means the targets we identify are more likely to be biologically relevant and safe for human trials.
What’s the difference between federated and centralized omics platforms?
Centralized platforms require you to upload and move data to a single location. This is not only slow (moving a petabyte of data can take weeks) but also creates massive security and compliance risks. Lifebit’s federated platform allows you to keep your data where it lives (e.g., in your own AWS, Azure, or Google Cloud bucket, or on-premise servers) while our AI “visits” the data to perform analysis. This “data visiting” model is the only way to collaborate globally while adhering to strict data residency laws like those in the EU or China.
Can Lifebit’s AI-powered omics analytics reduce clinical trial costs?
Absolutely. The most expensive part of a trial is failure. By using virtual clinical simulations and digital patient models, we can identify which patient subgroups are most likely to respond to a drug before the trial even starts. This allows for “enrichment” strategies, where you only recruit patients with the specific molecular signature the drug targets. This reduces the “all-comers” risk, requires fewer total patients to achieve statistical significance, and focuses resources on the most promising leads.
How does AI handle the complexity of multi-omics integration?
Integrating different layers of biology (DNA, RNA, Protein) is mathematically challenging because the data types are so different. AI-powered omics analytics uses “late fusion” or “intermediate fusion” techniques where each data type is first processed by a specialized sub-network before being combined into a single predictive model. This allows the AI to capture the unique information in each layer while still understanding the holistic state of the cell.
Is this technology applicable to rare diseases?
Yes, and it is often the only hope for rare disease patients. Because rare disease data is, by definition, scarce, federated AI is essential. It allows researchers to aggregate data from five patients in one country and ten in another to build a large enough cohort for meaningful analysis, all without violating the privacy of these vulnerable populations.
End Trial-and-Error: Scale Your AI-Powered Omics Analytics Today
The era of trial-and-error drug discovery is ending. We are entering the age of “In Silico First,” where the majority of the discovery work happens in a digital environment before a single drop of liquid is moved in a wet lab. AI-powered omics analytics is no longer a luxury for the top five pharma companies; it is the essential infrastructure of modern medicine.
By adopting a federated approach, prioritizing data quality, and using explainable AI models, your organization can move from being data-rich to being insight-driven. The goal is not just to collect data, but to transform that data into life-saving treatments.
At Lifebit, we are committed to providing the tools—from the Trusted Research Environment to advanced AI for genomics—that make this transition possible. We believe that the next great medical breakthrough shouldn’t be delayed by a data silo or a slow server. The future of precision medicine is federated, collaborative, and powered by AI. Are you ready to lead the way?
Learn more about GA4GH open standards and how they power our global research networks.

