Cracking the Genetic Code with Machine Learning

AI for Genomics: Cut Sequencing Analysis from 16 Hours to 5 Minutes
AI for genomics is fundamentally changing how researchers analyze genetic data, diagnose diseases, and develop personalized treatments. By moving beyond traditional statistical models and embracing deep learning, the field is undergoing a shift from descriptive biology to predictive, digital biology. Here’s what it does:
What AI for Genomics Delivers:
- Faster variant calling — Deep learning models like DeepVariant and Clair3 identify genetic mutations with higher accuracy than traditional methods by treating genomic signals as image-like patterns.
- Scalable sequencing analysis — GPU acceleration reduces analysis time from 16 hours to under 5 minutes, enabling high-throughput clinical workflows.
- Precision medicine — AI predicts disease risk and personalizes treatments by analyzing multi-omics data, including transcriptomics and proteomics, to see the full biological picture.
- Rare disease diagnosis — Machine learning pinpoints causative mutations from whole-genome sequencing in hours, not months, ending the “diagnostic odyssey” for many families.
- CRISPR optimization — AI predicts off-target effects and designs safer gene edits by modeling the complex interactions between guide RNAs and the genome.
- Data integration — Unsupervised learning finds hidden patterns across massive, siloed datasets that are too large for human researchers to manually curate.
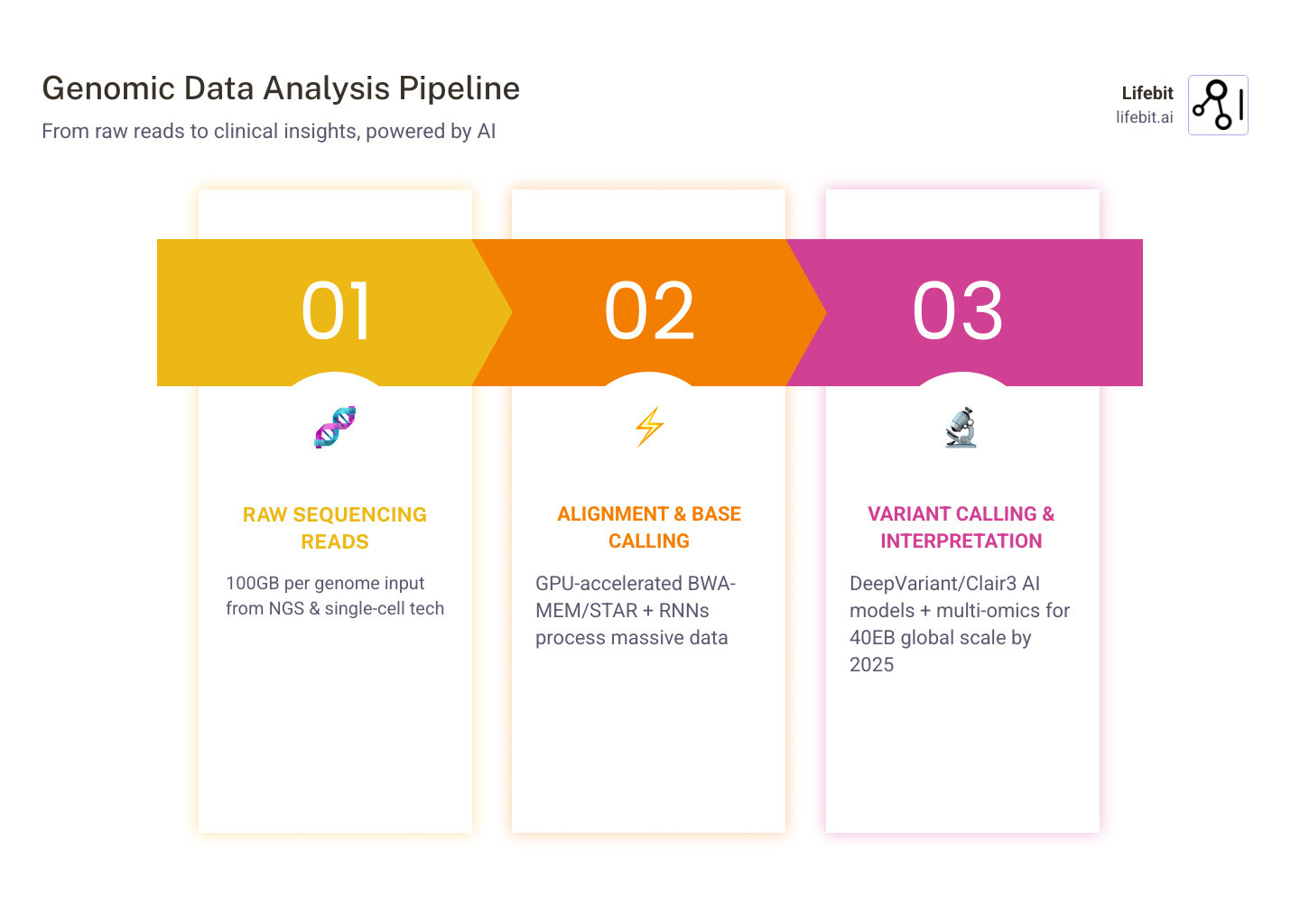
Sequencing a single human genome generates roughly 100 gigabytes of raw data—more than doubling after analysis. By 2025, storing all human genome data will require an estimated 40 exabytes, equivalent to 8 times more storage than every word spoken in history. This data explosion is driven by the falling cost of sequencing, which has outpaced Moore’s Law for over a decade.
Traditional bioinformatics tools, originally designed for a different era of computing, simply can’t keep up. The end of Moore’s Law means we need new computing paradigms—specifically accelerated computing and AI—to handle this data explosion. That’s where AI steps in, transforming raw biological noise into actionable clinical insights.
The genomics data bottleneck is real:
- High-throughput sequencing technologies like next-generation sequencing (NGS) and single-cell sequencing create massive, multi-dimensional datasets that require petabytes of temporary storage during processing.
- Only 2% of the human genome is coding—the remaining 98%, often called “dark DNA,” holds critical regulatory information that’s poorly understood but increasingly linked to complex diseases like diabetes and heart disease.
- Variants of uncertain significance (VUS) create diagnostic bottlenecks that delay clinical decisions; AI helps classify these variants by predicting their impact on protein folding and function.
- Most genomic datasets are siloed across institutions due to privacy concerns, making large-scale analysis nearly impossible without advanced technologies like federated learning.
AI doesn’t just speed things up. It changes what’s possible. Machine learning models find patterns in unstructured genomic data that humans and traditional algorithms miss entirely. Deep learning treats DNA sequences like natural language, predicting gene functions and disease risks with unprecedented accuracy. This is the era of “Foundation Models” for biology, where AI is trained on the entire known universe of genetic sequences to understand the grammar of life.
From base calling during sequencing to identifying cancer mutations for targeted therapies, AI now powers every stage of the genomics pipeline. Tools like NVIDIA Parabricks accelerate secondary analysis up to 80x, while models like AlphaFold and AlphaMissense predict protein structures and variant effects across entire proteomes. These tools are not just incremental improvements; they are the infrastructure for a new age of medicine.
But this revolution comes with serious challenges. Algorithmic bias, data privacy risks, and the lack of regulatory frameworks threaten to create new health inequities even as AI opens doors to precision medicine. Ensuring that AI models are trained on diverse populations is critical to preventing the “genomic divide.”
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, a federated data platform that enables secure, compliant AI for genomics across global healthcare systems. With over 15 years in computational biology, bioinformatics, and health-tech entrepreneurship, I’ve contributed to tools like Nextflow and worked at the Centre for Genomic Regulation to build systems that power precision medicine and biomedical data integration at scale.
This guide breaks down how AI is cracking the genetic code—from variant calling and GPU acceleration to multi-omics integration and federated learning—and what it means for the future of medicine.
AI for genomics terms to remember:
Stop Sequencing Bottlenecks: How AI for Genomics Hits 99% Accuracy
At its core, AI for genomics is about pattern recognition at a scale that exceeds human cognition. When we sequence a genome, we aren’t just reading a book; we are trying to assemble millions of tiny fragments of a puzzle while looking for microscopic typos. This process, known as variant calling, is where AI has made its most immediate and profound impact.
Traditional statistical methods often struggle with the “noise” inherent in sequencing data, such as PCR artifacts or chemical signal fluctuations. Modern genomic workflows now use deep learning models like DeepVariant (developed by Google) and Clair3. These tools treat genomic data almost like an image, using Convolutional Neural Networks (CNNs) to distinguish between actual genetic mutations and simple sequencing errors. By training on “truth sets” of highly validated genomes, these models have achieved accuracy levels that surpass traditional heuristic-based callers.
According to research on the transformative role of Artificial Intelligence in genomics, AI excels at processing unstructured raw sequencing reads that traditional bioinformatics tools find overwhelming. To make sense of this data, we rely on gold-standard alignment algorithms that have been re-engineered for the AI era:
- BWA-MEM: A leading algorithm for mapping DNA sequence reads to a reference genome. More info on BWA-MEM.
- STAR: The ultrafast standard for RNA-seq data alignment, helping us understand gene expression and how it changes in response to disease. More info on STAR.

GPU Acceleration and the End of Moore’s Law
For decades, we relied on CPU power getting faster every year. But as the volume of next-generation-sequencing data exploded, we hit a wall. Moore’s Law—the observation that transistors on a chip double every two years—is no longer enough to keep pace with the 40-exabyte data mountain we are building. This has led to the rise of “Huang’s Law,” which suggests that GPU performance for AI will more than double every two years.
The solution? GPU acceleration. By using thousands of small cores to perform calculations in parallel, we can accelerate secondary analysis software by up to 80x. This is critical for clinical applications where time-to-result can be the difference between life and death.
Consider these staggering statistics that highlight the shift:
- NVIDIA Parabricks can reduce the runtime of a germline HaplotypeCaller from 16 hours on a CPU to less than 5 minutes using GPU acceleration. This allows a single server to process dozens of genomes per day instead of just one.
- The Smith-Waterman alignment algorithm, a cornerstone of bioinformatics, is accelerated 35x on the NVIDIA H100 Tensor Core GPU.
- New systems like the PacBio Revio, powered by NVIDIA GPUs, can sequence high-accuracy long reads at scale for under $1,000 per genome, providing a more complete picture of structural variants that short-read sequencing often misses.
- Companies like Ultima Genomics are pushing the envelope further, aiming for high-throughput whole genome sequencing at just $100 per sample, making population-scale genomics economically viable for the first time.
This isn’t just about saving time; it’s about accessibility and democratization. When analysis takes minutes instead of days and costs $100 instead of $10,000, whole-genome sequencing becomes a routine part of every patient’s check-up, rather than a last-resort diagnostic tool. This shift enables “preventative genomics,” where we identify risks before symptoms even manifest.
AI for Genomics: Diagnose Rare Diseases in Hours, Not Years
The real magic happens when we move from “reading” the genome to “interpreting” it. AI for genomics is the engine behind precision medicine, where treatments are custom-tailored to your specific genetic makeup. This is the transition from reactive medicine to proactive, personalized healthcare.
We are moving beyond a “one-size-fits-all” approach. By integrating multi-omics data—combining genomics with proteomics (proteins), transcriptomics (RNA), and epigenomics (chemical modifications to DNA)—AI can predict how a specific patient will respond to a specific drug. As noted in the evolution and future of medical artificial intelligence, we are transitioning from Large Language Models (LLMs) to agent-based systems that can steer complex medical data to suggest therapies. These agents can cross-reference a patient’s genetic variants with thousands of clinical trials and millions of research papers in seconds.
For families dealing with rare diseases, this is a lifeline. In the past, children with undiagnosed genetic disorders might spend years undergoing tests, a process known as the “diagnostic odyssey.” Today, AI-driven analysis of genomes can pinpoint causative mutations in a matter of hours. Seattle Children’s Hospital, for instance, has used high-throughput nanopore sequencing to diagnose critical-care newborns in record time, allowing for immediate surgical or pharmacological intervention.
Comparing Traditional vs. AI-Driven Precision Medicine
| Feature | Traditional Diagnostics | AI-Driven Precision Medicine |
|---|---|---|
| Analysis Speed | Days to Weeks | Minutes to Hours |
| Data Scope | Single Gene or Panel | Whole Genome + Multi-omics |
| Accuracy | Statistical Inference | Deep Learning Pattern Recognition |
| Treatment | Symptom-based | Genotype-targeted |
| Early Detection | Often after symptoms appear | Predictive via liquid biopsies |
| Drug Response | Trial and Error | Pharmacogenomic Prediction |
AI for Genomics in Cancer Care and CRISPR Editing
In oncology, AI is a game-changer. We are now using AI to analyze liquid biopsies—blood tests that detect circulating tumor DNA (ctDNA). This allows for the detection of Molecular Residual Disease (MRD), catching cancer recurrence months before a scan would show a physical tumor. AI models are trained to distinguish the tiny fraction of tumor DNA from the vast background of healthy cell-free DNA in the blood.
Two of the most significant breakthroughs in this space are:
- AlphaFold: This AI system by Google DeepMind achieved highly accurate protein structure prediction, solving a 50-year-old biological challenge. It helps us understand how mutations change protein shapes, leading to disease. The latest version, AlphaFold 3, can even model how DNA, RNA, and ligands interact with proteins, accelerating drug discovery.
- AlphaMissense: This tool predicts the effect of missense variants across the entire human proteome, helping clinicians decide if a “variant of uncertain significance” is actually pathogenic. This reduces the ambiguity in clinical reports and provides clearer answers to patients.
AI is also making CRISPR-Cas9 safer. By predicting “off-target” effects—where the gene-editing tool might accidentally cut the wrong part of the DNA—machine learning models help researchers design guide RNAs that only edit the intended part of the genomics sequence. This is essential for moving gene therapies into human clinical trials, where safety is the paramount concern. AI models like CRISPR-Net and DeepCRISPR use deep learning to model the thermodynamics and kinetics of DNA binding, providing a level of precision that was previously impossible.
Solve the 40-Exabyte Crisis: Secure AI for Genomics Without Data Leaks
We are facing a data deluge that threatens to overwhelm our current infrastructure. If an estimated 40 exabytes will be required to store human genome data by 2025, we have to ask: where does it go, and who can see it? The sheer volume of data makes traditional cloud storage and transfer methods both expensive and risky.
The challenges are not just technical, but ethical and regulatory:
- Data Privacy: Genetic data is the ultimate identifier. Unlike a credit card number, you can’t change your DNA if it’s leaked. This makes genomic data a high-value target for cyberattacks and necessitates military-grade security.
- Algorithmic Bias: If an AI is trained primarily on genomes from one ethnic group (currently, over 80% of genomic data comes from individuals of European ancestry), it may be less accurate for others. We must ensure equitable access and diverse datasets to prevent health disparities from being baked into AI models.
- Computational Demands: The energy required to process omics data at scale is massive. Moving toward more efficient GPU-accelerated workflows is not just a speed requirement, but a sustainability one.
- Data Sovereignty: Many countries now have strict laws (like GDPR in Europe or the Data Security Law in China) that prevent genomic data from leaving national borders. This creates a massive hurdle for international research collaboration.
At Lifebit, we address these problems through a federated AI platform. Instead of moving sensitive data to the AI (which creates security risks and incurs massive egress costs), we bring the AI to the data. This is known as “data visiting.” This allows for secure, real-time access to global biomedical data while maintaining strict genomic data privacy and compliance.
Our approach utilizes Trusted Research Environments (TREs), which are secure spaces where researchers can analyze data without ever being able to download the raw, identifiable files. By using federated learning, we can train AI models across multiple institutions simultaneously. The model learns from the data at each site, but only the “insights” (the mathematical weights of the model) are shared and aggregated. This ensures that the underlying patient data never moves, satisfying both security experts and national regulators. This is the only way to reach the scale needed for the 40-exabyte future while keeping patient trust intact.
Beyond Labels: How AI for Genomics Decodes 98% of Dark DNA
The next frontier in AI for genomics is unsupervised learning and the rise of biological foundation models. Most current models are “supervised,” meaning they learn from data that humans have already labeled (e.g., “this mutation causes this disease”). But the 98% of our genome that is “non-coding” doesn’t come with labels. It is a vast, mysterious landscape of regulatory switches, enhancers, and silencers.
Emerging models like GenomeOcean and Evo are designed to learn the “natural language” of genomes. Just as ChatGPT predicts the next word in a sentence by understanding the context of millions of books, these models read vast amounts of genomic sequences to uncover hidden patterns and relationships within DNA. They learn the “grammar” of the genome—how certain sequences interact over long distances to turn genes on or off.
According to recent scientific research on GenomeOcean, this approach allows AI to “autocomplete” missing genetic code and predict the functional impact of mutations in non-coding regions. This is invaluable for synthetic biology applications, such as designing new biofuels, carbon-sequestering plants, or novel pharmaceuticals that target the regulatory machinery of a cell rather than just the proteins themselves.
Scaling AI for Genomics with Federated Governance
As we move toward 2030, the focus will shift from simple data analysis to federated governance. We cannot solve the world’s biggest health challenges—like cancer, Alzheimer’s, or pandemic preparedness—by working in isolation. We need a way for researchers in New York, London, Singapore, and beyond to collaborate without ever compromising patient privacy or institutional intellectual property.
Lifebit’s Trusted Research Environment (TRE) and Trusted Data Lakehouse (TDL) provide this foundation. By using federated learning, we can train AI models on diverse global datasets—improving accuracy and reducing bias—all while the data stays safely behind the firewalls of the original hospitals or biobanks. This “collaborative AI” model is the key to unlocking the secrets of rare diseases that may only have a handful of patients in any single country, but hundreds when viewed globally.
Furthermore, generative AI is now being used to create synthetic genomic data. This synthetic data mimics the statistical properties of real patient data but contains no actual identifiable information. This allows researchers to develop and test AI models in “sandbox” environments before deploying them on real, sensitive datasets, further accelerating the pace of innovation while minimizing risk.
AI for Genomics: 3 Critical Questions Every Researcher Must Answer
How does AI improve the accuracy of genome sequencing compared to traditional bioinformatics?
Deep learning models like DeepVariant use convolutional neural networks (CNNs) to treat genomic signals as images. Traditional methods rely on hand-crafted heuristics and statistical thresholds that often fail to distinguish between true variants and systematic sequencing errors (noise). By “seeing” the data in its raw form, AI can identify subtle patterns that indicate a real mutation, significantly reducing false positives and false negatives. This is especially true in “difficult-to-map” regions of the genome where traditional math-based callers often struggle.
What are the biggest ethical and regulatory risks in genomic AI today?
The primary risks include algorithmic bias—where models are trained on non-diverse datasets, leading to lower diagnostic accuracy for underrepresented populations—and the potential for data breaches involving highly sensitive, identifiable genetic information. There is also the concern of “genetic discrimination” in insurance or employment if data is not properly governed. Furthermore, the “black box” nature of some deep learning models makes it difficult for clinicians to understand why an AI made a specific prediction, which is a major hurdle for regulatory approval and clinical trust.
Can AI predict rare diseases before symptoms appear, and how?
Yes, by integrating multi-omics data and identifying subtle patterns in non-coding regions of the genome, AI can flag predispositions for rare diseases years before clinical manifestation. For example, AI can analyze transcriptomic signatures (how genes are being expressed) to find early signs of metabolic distress or cellular dysfunction. This is particularly effective in identifying metabolic disorders or predispositions to childhood-onset conditions, allowing for early interventions like dietary changes or gene therapies that can prevent permanent organ damage or developmental delays.
Start Your AI for Genomics Journey: Secure, Scalable, and Compliant
The integration of AI for genomics is no longer a futuristic concept—it is the current reality of modern medicine and the only viable path forward for digital biology. From accelerating analysis with NVIDIA Parabricks to predicting protein folding with AlphaFold, AI is the only tool powerful enough to help us navigate and make sense of the 40-exabyte future.
As we look toward the next decade, the convergence of AI, cloud computing, and high-throughput sequencing will make “genomic intelligence” a standard part of the healthcare ecosystem. We will see the rise of Digital Twins, where a patient’s genomic and clinical data are used to create a virtual model to test drug responses before they are administered in real life. We will see the total eradication of certain genetic diseases through AI-guided CRISPR therapies. And we will see a global network of federated data that allows for real-time disease surveillance and drug discovery.
However, the power of AI must be balanced with responsibility. Strategic integration requires secure collaboration, bias mitigation, and robust regulatory frameworks. At Lifebit, we are committed to providing the best ai genomics guide and the infrastructure needed to power this revolution safely. Our mission is to ensure that the benefits of genomic AI are available to everyone, regardless of where they live or what their genetic heritage is.
Our federated AI platform ensures that the next generation of medical breakthroughs is built on a foundation of security and trust. Whether you are a researcher in the USA, a clinician in Europe, or a policymaker in Singapore, the tools to crack the genetic code are now within reach. The 40-exabyte challenge is not a crisis, but an opportunity to redefine what it means to be healthy.
Get started with accelerated genomic analysis today and join us in shaping the future of precision medicine.

