Unlocking Insights: What a Bioinformatics Platform Can Do For You
Understanding Bioinformatics Platforms: Your Gateway to Modern Data-Driven Research
A bioinformatics platform is a unified computational environment that integrates data management, workflow orchestration, analysis tools, and collaboration features to accelerate biological research. Unlike standalone tools or traditional HPC clusters, these platforms provide end-to-end solutions for processing, analyzing, and sharing complex biological datasets, forming the operational backbone for modern life sciences organizations.
Key components of a bioinformatics platform include:
- Data Management: This goes beyond simple storage. It involves automated ingestion of raw data (e.g., FASTQ, BCL files), running standardized quality control checks (like FastQC), and capturing rich, structured metadata that adheres to FAIR principles (Findable, Accessible, Interoperable, Reusable). This ensures every dataset is discoverable and its context is understood.
- Workflow Orchestration: This is the engine that drives analysis. It allows for the execution of complex, multi-step bioinformatics pipelines in a standardized, reproducible, and scalable manner. It includes version control for both pipelines (e.g., via Git integration) and software dependencies (via containers like Docker or Singularity), ensuring that an analysis run today can be perfectly replicated years later.
- Analysis Environments: Research doesn’t end with a pipeline. This component provides interactive spaces like Jupyter notebooks and RStudio, integrated with the platform’s data and compute resources. It also includes visualization tools, such as integrated genome browsers (IGV) and custom dashboards, allowing researchers to explore results, test hypotheses, and generate publication-quality figures.
- Security & Governance: For handling sensitive patient or proprietary data, this is non-negotiable. It encompasses granular Role-Based Access Controls (RBAC), comprehensive audit trails logging every action, and robust compliance frameworks to meet standards like HIPAA and GDPR.
- Collaboration: Science is a team sport. Platforms facilitate this with secure project workspaces where teams can share data, pipelines, and results. Permissions can be finely tuned to control who can view, edit, or execute analyses, enabling seamless collaboration both within and between organizations.
The life sciences industry is experiencing an unprecedented data explosion. With genomics data doubling every seven months and over 105 petabytes of precision health data managed on modern platforms, the scale is immense. Add to this the 2.43 million monthly workflow runs executed globally, and it’s clear that researchers must turn this data deluge into actionable insights.
Traditional approaches, like using standalone tools or managing on-premise HPC clusters, create severe bottlenecks. An analyst might spend days installing software and managing dependencies, only to find their workflow isn’t portable to a collaborator’s system. Data gets siloed in individual hard drives or disparate servers, moved insecurely via FTP, making integrated analysis nearly impossible. Collaboration becomes a manual, error-prone process of emailing scripts and results, and ensuring compliance is a constant struggle. Modern bioinformatics platforms solve these problems by creating a single pane of glass for your entire research ecosystem. They centralize data, standardize workflows, and enable secure collaboration while scaling from pilot studies to population-level analyses.
Unlike LIMS or ELNs, which focus on sample and lab process tracking (the pre-analytical phase), a bioinformatics platform is designed for the post-analytical world of large-scale data interpretation. It integrates data governance, workflow management, and advanced analytics, going far beyond the capabilities of traditional HPC clusters, which provide raw compute but lack the integrated management and governance layers.
As Maria Chatzou Dunford, CEO and Co-founder of Lifebit, I’ve spent over 15 years building computational biology tools and bioinformatics platforms that power precision medicine research globally. My experience developing Nextflow and leading federated genomics initiatives has shown me how the right platform architecture can transform research outcomes.
Key terms for bioinformatics platform:
Core Capabilities of a Modern Bioinformatics Platform
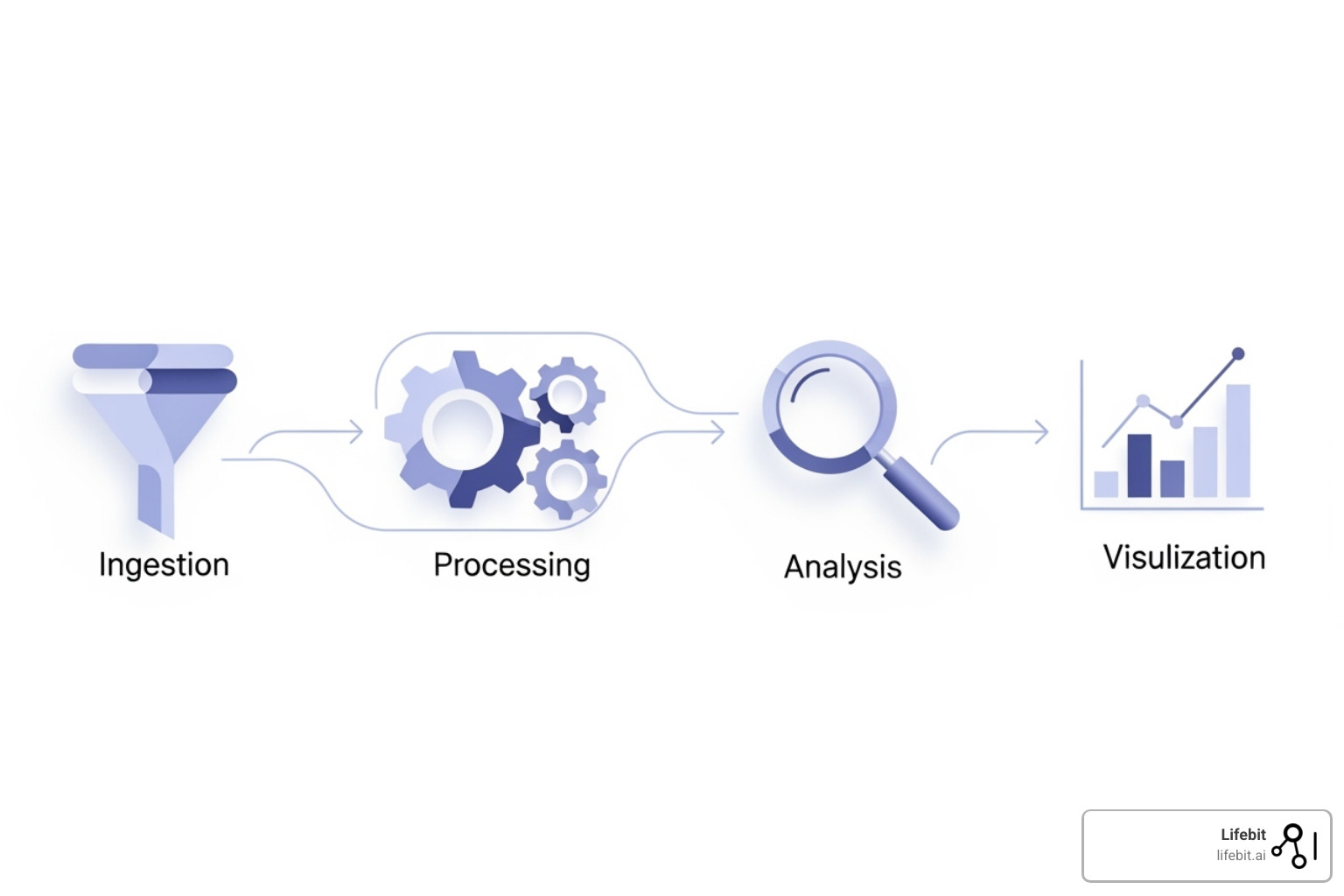
A modern bioinformatics platform acts as a research command center, providing a “single pane of glass” view of the entire research ecosystem. Upon arrival, data is automatically ingested, with metadata captured to ensure full data provenance. Automation is a key differentiator; for instance, new sequencing data appearing in a designated storage bucket can trigger quality control pipelines automatically, generating a MultiQC report for review, as seen with advanced analytics with Nextflow pipelines. This ensures workflows are portable and not locked into a single system or cloud provider.
Unifying Data, Pipelines, and Compute
A robust bioinformatics platform streamlines the handling of complex biological data. Data ingestion automation consistently processes diverse formats like FASTQ, BAM/CRAM, VCF, and next-generation formats like HDF5/Zarr with end-to-end quality control. Beyond genomics, true multi-modal data support is critical. For example, in oncology research, a platform can integrate a patient’s WGS data to find driver mutations, RNA-seq data to see their expression, proteomics data to confirm protein levels, and clinical imaging data to assess tumor morphology—all linked to treatment response outcomes. This holistic view is where novel discoveries are made.
The platform provides a pipeline catalog of validated, version-controlled workflows (e.g., from communities like nf-core) and allows users to bring-your-own-tools via containerization (e.g., Docker, Singularity). This flexibility, central to a distributed data analysis platform, ensures data and tools work together following FAIR principles (Findable, Accessible, Interoperable, and Reusable). It turns a chaotic collection of files and scripts into a structured, queryable research asset.
Workflow and Compute Orchestration
A bioinformatics platform orchestrates computation at any scale. It supports powerful workflow languages like Nextflow, which excels at defining complex, scalable pipelines through its reactive dataflow paradigm that simplifies parallelization and error handling. Advanced schedulers and orchestrators like Kubernetes are used under the hood to dynamically allocate resources and manage execution across diverse environments. This includes major cloud providers (AWS, GCP, Azure) and on-premise systems (like Slurm or LSF clusters) with various storage types (POSIX filesystems, object storage). This hybrid approach enables bringing computation to the data, a core principle of modern workflows and distributed computing for biomedical data, maximizing efficiency and security.
Ensuring Scientific Reproducibility and Portability
Modern bioinformatics platforms make reproducibility automatic, not an afterthought. Pipelines and reference genomes are versioned and pinned to specific runs, while detailed run tracking and lineage graphs provide a complete, immutable audit trail. This lineage graph is a powerful feature, capturing every detail: the exact container image used (e.g., nf-core/rnaseq:3.10.1), the specific parameters chosen (--aligner star), the reference genome build (GRCh38), and the checksums of all input and output files. This creates an unbreakable chain of provenance, essential for publications, patents, and regulatory filings. Container images ensure consistent software environments, eliminating the “it works on my machine” problem entirely. A commitment to open standards and clear export paths prevents vendor lock-in, a transparency that extends to interactive environments like reproducibility in life sciences: CloudOS + Jupyter collaboration. Even no-code interfaces are built on inspectable code, ensuring users maintain full scientific control and transparency.
Powering Findy: From Genomics to AI-Driven Medicine
A bioinformatics platform is a powerful tool for scientific and clinical research, designed to improve time-to-results, boost throughput, and reduce costs. By combining standardized workflows, such as standardised bioinformatics pipelines, with advanced analytics, modern platforms push the boundaries of biological research.
Foundational ‘Omics’ and Multi-omics Analysis
NGS secondary analysis forms the backbone of genomic research, with battle-tested pipelines for WGS, WES, and RNA-seq. Single-cell analysis workflows uncover cellular heterogeneity using single-cell RNA-seq and ATAC-seq. Platforms also support epigenomics (e.g., ChIP-seq, bisulfite sequencing) to study gene regulation. Comprehensive metagenomics support, from metabarcoding to shotgun metagenomics, is also crucial, backed by our expertise in genomics and metagenomics research. Proteomics capabilities include advanced protein structure prediction (e.g., AlphaFold), functional annotation, and interaction analysis. The platform’s true power lies in multi-omics integration. For instance, in an Alzheimer’s study, researchers can integrate genomics (to identify APOE risk variants), transcriptomics (to quantify neuroinflammatory gene expression), and proteomics (to measure amyloid-beta levels in CSF), and correlate these molecular signatures with clinical cognitive scores in a unified cohort browser to reveal novel disease mechanisms and biomarkers.
Interactive Analysis and Clinical Translation
While pipelines automate heavy lifting, interactive exploration in environments like JupyterLab and RStudio is crucial for custom analyses, hypothesis testing, and visualization. For non-programmers, intuitive no-code dashboards and cohort browsers make complex data accessible. The platform bridges research and clinical care by supporting assay development and validation with structured, GxP-compliant workflows. For example, a platform can provide a locked-down environment to validate a new diagnostic panel, automatically running it against reference samples, generating performance reports on sensitivity and specificity, and using e-signatures to formally approve each stage. Clinical report generation is automated, integrating with LIMS and maintaining full traceability and change control with immutable audit trails. This ensures compliance with standards like 21 CFR Part 11, which is critical for data analysis in Trusted Research Environments.
Accelerating AI/ML in Research
A modern bioinformatics platform provides comprehensive support for AI/ML workflows. It ensures secure data access for AI model training without compromising privacy, often through federated learning techniques. This is paired with robust MLOps practices for model governance, including experiment tracking (e.g., with MLflow), model versioning in a central registry, and deployment for inference. For example, a platform can host a trained deep learning model that predicts drug response from gene expression data, allowing new patient samples to be analyzed in near real-time. AI assistants and copilots, like our Accelerate Nextflow with NF Copilot feature, help researchers build and optimize pipelines more efficiently. The results are tangible: teams often see correlations of 70-93% between predicted and experimental data, leading to a 60% or greater reduction in required wet lab experiments.
Secure and Scalable Operations: Deployment, Governance, and Cost Management

A scalable bioinformatics platform must operate securely, cost-effectively, and in full compliance with industry regulations. It needs to manage sensitive patient data and facilitate cross-institutional research while maintaining rigorous audit trails, a challenge addressed in our Lifebit approach to data governance & security (White paper).
Flexible Deployment and Data Management
A modern platform offers flexible deployment models: on-premises for maximum control, cloud for scalability, and hybrid approaches for the best of both. This flexibility is powered by federated analysis, as exemplified by the Lifebit Federated Biomedical Data Platform. Instead of moving data, this approach brings the analysis to the data. Technically, the platform sends a containerized analytical workflow to a secure ‘enclave’ within the data custodian’s environment. The workflow executes locally, and only the aggregated, non-identifiable results (e.g., p-values, model parameters) are returned to the researcher. This model is used by large-scale initiatives like the UK Biobank’s Research Analysis Platform to enable secure research while respecting data residency. Effective data lifecycle management automatically transitions data through active, archival, and cold storage tiers to optimize costs. This is all part of our Trusted Data Lakehouse concept, which brings analysis capabilities to controlled datasets.
Enterprise-Grade Security and Compliance for your bioinformatics platform
Security is fundamental to any bioinformatics platform. Key features include end-to-end encryption, Role-Based Access Control (RBAC), and comprehensive audit logs. Specialized PHI and PII protection with data masking and secure key management maintains patient trust. Our platforms meet numerous compliance standards, including HIPAA, GDPR, SOC 2, ISO 27001/27701, FedRAMP, GxP (including 21 CFR Part 11), and CLIA/CAP. For HIPAA, this means implementing technical safeguards like encrypting all data at rest and in transit, enforcing unique user IDs, and maintaining immutable logs of all data access. For GxP, the platform must support formal validation (IQ/OQ/PQ), enforce strict change control, and ensure data integrity with features like e-signatures. These certifications underscore our commitment to operating secure Trusted Research Environments (TREs) for sensitive research.
Collaboration and Cost Optimization
The platform fosters collaboration through secure workspaces and projects that enable cross-institutional sharing and the creation of publishable narratives. Cost management is critical at scale. The platform provides cost estimators, budgets, and quotas to prevent overspending. Costs are automatically optimized via autoscaling, use of spot/preemptible instances, and automated storage tiering. For example, a large-scale WGS analysis project might cost over $25,000 using standard on-demand cloud instances. By intelligently leveraging spot instances, which are up to 90% cheaper, and automatically re-running any interrupted jobs, a smart platform can reduce the total compute cost to under $5,000. Egress controls, detailed run dashboards, and proactive alerts help monitor performance and costs. These capabilities operate under our Trusted Operational Governance (Airlock) framework, ensuring security and compliance are never compromised.
Choosing and Implementing the Right Platform for Your Organization
Choosing the right bioinformatics platform is a long-term strategic decision. It’s about finding an environment that fits your team, workflows, and vision for data-driven research. Successful implementation requires careful planning, focusing on migration, onboarding, and change management, all while prioritizing the key features of a Trusted Research Environment.
What to Look For in a Bioinformatics Platform
When evaluating platforms, look for real-world performance benchmarks and case studies. Analyze the Total Cost of Ownership (TCO), not just the sticker price, and ensure pricing models align with your usage patterns (e.g., per-user, per-analysis, or resource-based). Crucial integration capabilities include REST APIs, SDKs, and SSO support for connecting with existing LIMS/ELN systems. Assess the platform’s scalability and elasticity—can it handle 10 samples as efficiently as 10,000? The user experience (UX) is also paramount; the platform should be intuitive for biologists with no-code interfaces, yet powerful enough for bioinformaticians who need CLI access. Also, assess the quality of training, documentation, and support, including SLAs. Finally, consider the platform’s community and ecosystem, as open science principles and access to open-source pipelines, like those found on open science platforms for biology, provide long-term value.
The Path to Success: Migration and Onboarding
A methodical migration starts with a pilot design to validate performance on a representative project. Plan your data transfer strategy and allow time for pipeline porting and validation. Effective change management is critical for adoption. A common hurdle is resistance from researchers accustomed to their bespoke scripts. Overcome this by identifying ‘power users’ to act as internal champions, providing comprehensive hands-on training, and clearly demonstrating the platform’s value in reducing manual effort. While minimal IT requirements are common, coordinate with your IT team early. Typical timelines for initial production runs are 2-4 weeks. Measure success with KPIs like time-to-result, cost-per-run, user adoption rates, and even the number of new cross-departmental collaborations enabled by the platform.
Addressing Ethical, Legal, and Privacy Considerations
A good bioinformatics platform helps manage the responsibilities of using patient data. Consent management tools track data usage permissions; modern platforms can even support dynamic consent, where participants digitally manage their preferences over time. Data privacy by design is essential, incorporating automatic de-identification and data masking. The platform should help navigate legal frameworks like HIPAA and GDPR and simplify reporting for ethical review boards (IRBs). It can streamline interactions with Data Access Committees (DACs) by providing a secure portal for researchers to submit applications, for DAC members to review requests against consent permissions, and to automatically provision time-limited access once approved, all with a full audit trail. Advanced anonymization techniques enable large-scale analysis while protecting individual privacy, which is crucial for accessing controlled datasets from sources like dbGaP and biobanks.
Frequently Asked Questions about Bioinformatics Platforms
Here are answers to some of the most common questions about transitioning to a modern bioinformatics platform.
What is the main difference between a bioinformatics platform and using standalone tools on a cluster?
A bioinformatics platform integrates data management, workflow orchestration, security, and collaboration into a single, managed ecosystem. In contrast, using standalone tools on a cluster requires manual data movement, custom scripting to connect processes, and significant effort to ensure reproducibility and governance. The platform provides the automation and intelligence that allows researchers to focus on science instead of infrastructure.
How does a bioinformatics platform support both bioinformaticians and bench scientists?
Modern platforms cater to both user groups. For bioinformaticians, they offer powerful command-line interfaces, APIs, and SDKs to build and automate custom workflows. For bench scientists and clinicians, they provide intuitive graphical interfaces and no-code tools that allow them to run complex analyses and visualize results without programming. This dual approach breaks down barriers and fosters collaboration between computational and experimental teams.
Can a bioinformatics platform run in a hybrid environment (cloud and on-premise)?
Yes. Modern bioinformatics platforms are designed for hybrid environments, orchestrating workflows across on-premise HPC clusters and public clouds (AWS, Azure, GCP). They use a federated approach, bringing computation to the data’s location. This respects data residency requirements, optimizes costs, and allows organizations to leverage existing infrastructure while accessing the scalability of the cloud. The platform manages this complexity, making it feel like a single, seamless environment.
Conclusion: The Future of Data-Driven Biology is Integrated
The future of biological research lies in integrated, intelligent platforms that replace fragmented tools and workflows. A modern bioinformatics platform transforms how you work with data, accelerating research and enabling precision medicine at scale. By automating pipelines, harmonizing multi-omics data, and allowing AI models to securely access datasets, these platforms help researchers publish faster and translate findings to the clinic more efficiently.
The power of integration eliminates the friction that slows scientific progress, while the importance of federation allows for global collaboration while respecting data sovereignty and patient privacy. This approach is not just technically advanced; it’s ethically essential for the future of healthcare.
At Lifebit, our federated platform embodies this vision, bringing together our Trusted Research Environment (TRE), Trusted Data Lakehouse (TDL), and R.E.A.L. (Real-time Evidence & Analytics Layer) to amplify your research potential.
The next wave of breakthroughs will emerge from collaborative networks powered by integrated platforms. Ready to see how this integration can transform your research? Find how our platform can open up the full potential of your biomedical data by visiting Find the Lifebit Platform. The future of data-driven biology is here, and it’s waiting for you to be part of it.

