The Multi-Omics Marvel: A Platform for Every Insight

Stop Wasting Research: Find Targets 10x Faster with a Multi-Omics Data Analytics Platform
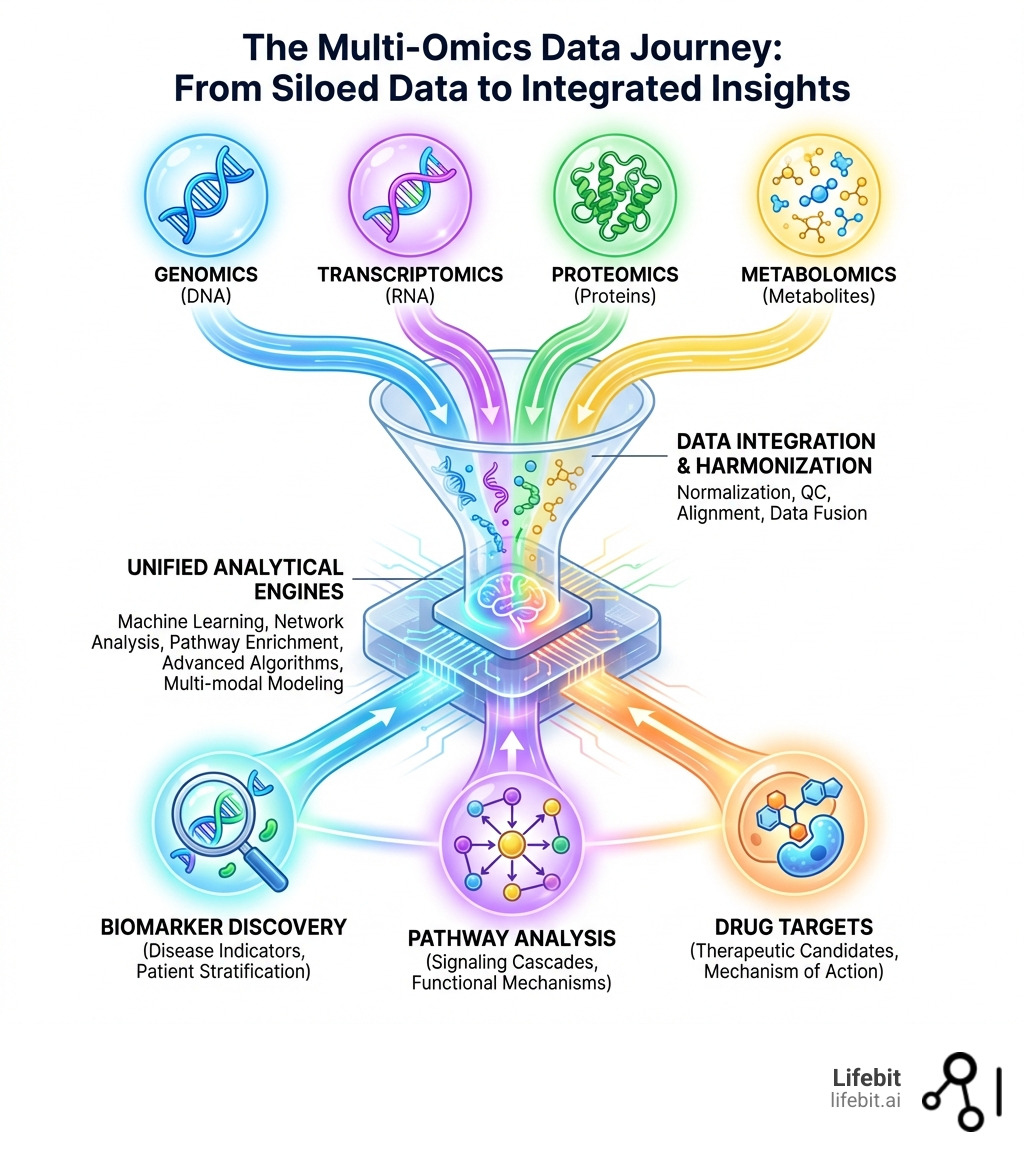
A multi-omics data analytics platform is a software solution that integrates and analyzes diverse biological data types—including genomics, transcriptomics, proteomics, and metabolomics—on a single scalable infrastructure. These platforms enable researchers to:
- Unify siloed datasets from multiple omics layers into one source of truth
- Apply advanced algorithms (machine learning, network analysis, pathway enrichment) to identify patterns across data types
- Visualize complex relationships through interactive dashboards and dimensionality reduction tools
- Accelerate biomarker findy and drug target identification by revealing cross-omics correlations
- Support users at every skill level, from biologists using no-code interfaces to bioinformaticians building custom workflows
Modern life sciences research generates massive volumes of interlinked data. A single study might produce genomic sequences, gene expression profiles, protein abundance measurements, and metabolite concentrations—all pointing to different pieces of the same biological puzzle. Yet most researchers analyze these datasets separately, missing the critical connections that only emerge when you look at the whole system.
The challenge isn’t just volume. It’s biological complexity. Genomics tells you what could happen. Transcriptomics shows what is happening at the RNA level. Proteomics reveals what proteins are actually doing. Metabolomics captures the end result—the small molecules that drive cell function. Each layer matters. But none tells the complete story alone.
That’s where integrated analysis changes everything. When you can correlate millions of data points across multiple omics layers, you uncover hidden relationships—the ligand-receptor pairs driving disease, the signaling pathways to target with drugs, the biomarkers that predict patient response. Studies using multi-omics platforms have identified predominant signaling pathways and therapeutic targets in colorectal cancer, finded meaningful cell-cell interactions in Parkinson’s disease, and revealed patterns that single-omics approaches simply cannot see.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, a pioneering genomics and biomedical data platform. Over 15 years building computational biology tools and contributing to frameworks like Nextflow, I’ve seen how the right multi-omics data analytics platform transforms raw data into clinical and research breakthroughs—when it’s built for security, scale, and real-world collaboration.
Common multi-omics data analytics platform vocab:
- bioinformatics platform
- What are the best biopharma data software solutions for large-scale research?
- clinical data integration platform
Why Single-Omics Is Killing Your Research—And How Integration Fixes It
We’ve all been there: staring at a beautiful heatmap of transcriptomic data, feeling like we’ve found the “smoking gun” for a disease, only to realize later that those RNA levels didn’t actually translate into functional proteins. This is the “single-omics trap.” When we analyze biological layers in isolation, we aren’t just missing a few details; we are often looking at a completely different story than what the biology is actually telling us.
The primary challenges driving the need for a multi-omics data analytics platform include:
- Data Silos: Research teams often store genomic data in one cloud bucket, proteomics in a local server, and clinical phenotypic data in a completely different database. Breaking these silos is the first step toward findy.
- Data Heterogeneity: A FASTQ file is nothing like a mass spec intensity table. Harmonizing these different formats so they “speak the same language” is a monumental task without an integrated platform.
- Scalability Issues: Modern datasets, especially single-cell and spatial omics, can involve hundreds of thousands of cells and millions of data points. Local computing infrastructure simply can’t keep up.
- Reproducibility: If a bioinformatician runs a custom script on their laptop, can another researcher reproduce those results six months later? Often, the answer is no.
The Problem with a Piecemeal Approach
When we take a piecemeal approach—analyzing one omics layer at a time—we suffer from a massive lack of biological context. It’s like trying to understand how a car works by only looking at the tires. You might see they are spinning, but you’ll never understand the engine, the fuel, or the driver.
This fragmented method leads to:
- Missed correlations: You might miss a crucial interaction where a specific genetic variant only causes disease when a particular metabolite is present.
- Inefficient workflows: Researchers spend 80% of their time cleaning and formatting data rather than actually analyzing it.
- Increased manual effort: Passing files back and forth between different specialized tools is a recipe for version-control nightmares.
The Power of a Unified View
In contrast, a unified multi-omics data analytics platform offers a holistic understanding. By establishing a “single source of truth,” we can move toward system-level biology. This isn’t just a buzzword; it’s the ability to see how a mutation in the DNA flows through the RNA, affects protein expression, and ultimately alters the metabolic state of a cell.
This unified view accelerates findy by allowing us to identify top ligand-receptor pairs and cell-cell interactions that are consistent across multiple experiments. It turns a “maybe” into a “statistically significant yes.”
Inside the Multi-Omics Data Analytics Platform: 4 Tools for Better Insights
What actually goes into a high-end multi-omics data analytics platform? It’s more than just a place to store files. It’s an ecosystem designed to take you from a raw sequencer run to a peer-reviewed insight.
The core components we build into these systems include:
- Data Ingestion: The ability to easily ingest, organize, and store omics, imaging, and real-world data (RWD).
- Data Harmonization: Automated pipelines that normalize data and remove “batch effects”—those annoying variations that happen just because samples were processed on different days or in different labs.
- Analytical Engines: The “brains” of the operation, where statistical models and machine learning algorithms live.
- Visualization Tools: Interactive dashboards that let you “play” with your data in real-time.
Seamless Data Integration: From Genomics to Metabolomics
To get a true “big picture” view, a platform must support an incredible diversity of data. This includes bulk and single-cell RNA-seq, proteomics (via mass spec or CITE-seq), epigenomics (like ATAC-seq), and even spatial transcriptomics, which preserves the tissue’s physical architecture.
But it doesn’t stop at omics. Modern research increasingly requires the integration of imaging data and phenotypic records. By breaking down the silos between these multimodal datasets, we can correlate a specific molecular signature with an MRI scan or a patient’s long-term clinical outcome.
The Analytical Powerhouse: Key Algorithms and Approaches
A multi-omics data analytics platform is only as good as the math behind it. We rely on industry-standard statistical algorithms for noise reduction and normalization to ensure the patterns we see are real biological signals, not technical artifacts.
Key approaches include:
- Machine Learning (ML): Algorithms like Extreme Gradient Boosting (XGBoost) are used for classification and predicting genotype-phenotype responses.
- Network Analysis: Tools like Similarity Network Fusion (SNF) help us see how different omics layers overlap and interact.
- Pathway Analysis: Identifying which biological pathways (like those found in KEGG) are significantly enriched across your datasets.
For those interested in the deep technical validation of these methods, the Scientific validation of web-based multi-omics integration provides a comprehensive look at how these suites function in a peer-reviewed context.
From Raw Data to Insight: Essential Visualization Features
Let’s be honest: nobody wants to look at a million-row Excel spreadsheet. We need to see the data.
Essential features for any researcher include:
- Dimensionality Reduction: Using PCA, UMAP, or t-SNE to turn complex, high-dimensional data into 2D or 3D scatterplots where clusters of similar samples become obvious.
- Interactive Heatmaps: Allowing you to zoom in on specific gene-protein correlations.
- Volcano Plots: Instantly identifying the most significantly up-regulated or down-regulated features.
2025 Guide: Choose a Multi-Omics Data Analytics Platform Without Security Risks
Choosing a multi-omics data analytics platform is a bit like choosing a house: you need to make sure the foundation is solid before you worry about the paint color. For us, the “foundation” is security and scalability.
| Feature | Standard Platform | Federated Platform (Lifebit) |
|---|---|---|
| Deployment Model | Cloud-based (SaaS) or On-Premise | Federated / Hybrid Cloud |
| Data Access | Data must be moved to the tool | Tool moves to the data (No movement) |
| User Interface | GUI or CLI | Unified No-code GUI + CLI |
| Security Model | Standard Encryption | Trusted Research Environments (TRE) |
| Pricing Model | Subscription / Seat-based | Scalable / Pay-per-use |
Deployment and Accessibility: Cloud vs. On-Premise
The old way of doing things—on-premise servers—is becoming a bottleneck. While they offer control, they lack the “burst” capacity needed for large-scale multi-omics projects. Most modern platforms are now cloud-based (SaaS), allowing for massive scalability.
However, the best solutions often use a hybrid cloud or federated model. This allows organizations to keep their most sensitive data behind their own firewall or in a private cloud configuration while still leveraging the power of cloud-based analytics. And because these tools are web-based, you should always ensure you are using Recommended browsers for web-based tools like Chrome or Firefox for the best performance, especially when rendering complex WebGL visualizations.
Security and Compliance: A Non-Negotiable Requirement
When you’re dealing with patient genomes, security isn’t just a “nice to have”—it’s a legal requirement. A robust multi-omics data analytics platform must conform to GDPR, HIPAA, and other global privacy regulations.
At Lifebit, we advocate for a federated architecture. Instead of moving massive, sensitive datasets into a central repository (which creates a huge security risk), we bring the analysis to the data. This is often done within a Trusted Research Environment (TRE), a secure space where researchers can analyze data without ever being able to download the raw, identifiable information. This “security-first” infrastructure ensures that data governance remains in the hands of the data owner.
Usability for Every Skill Level: Empowering Biologists and Bioinformaticians
One of the biggest problems in omics research is the “bioinformatics bottleneck.” There are never enough bioinformaticians to go around.
A modern platform must cater to two distinct groups:
- Biologists: They need an intuitive, no-code Graphical User Interface (GUI). They want to click a button and see a pathway analysis, not write 500 lines of R code.
- Bioinformaticians: They need a Command-line Interface (CLI) and the ability to build custom, reproducible workflows using languages like Nextflow or WDL.
By providing pre-built pipelines and a user-friendly workflow builder, we empower the people who designed the experiment to actually analyze the results.
Cut Years Off Drug Discovery: Identify Targets in Months with a Multi-Omics Data Analytics Platform
What does all this technical wizardry actually achieve? It saves lives and cuts years off the drug findy timeline. By looking at the “big picture,” researchers are identifying drug targets that were previously invisible.
Revolutionizing Drug Findy and Biomarker Identification
In drug findy, the goal is to find the right target for the right patient at the right time. Multi-omics platforms facilitate this through:
- Target Identification: Identifying proteins that are consistently overexpressed across different patient cohorts.
- Genotype-Phenotype Prediction: Understanding how a specific genetic makeup will respond to a new therapeutic.
- Patient Stratification: Grouping patients based on their molecular signatures to ensure clinical trials are more likely to succeed.
Case Study: Colorectal Cancer
Researchers used integrated multi-omics to identify predominant signaling pathways and potential biomarkers across multiple experiments. By combining transcriptomic and proteomic data, they identified therapeutic targets that would have been missed if they only looked at DNA mutations.
Case Study: Parkinson’s Disease
A meta-analysis of Parkinson’s data—integrating transcriptomics, proteomics, and metabolomics—unveiled meaningful cell-cell interactions and ligand-receptor pairs. This allowed researchers to prioritize drug targets that specifically address the neurodegenerative pathways involved.
The Proof is in the Publications: Scientific Validation
We don’t just take a platform’s word for it; we look at the peer-reviewed evidence. Scientific validation is critical for ensuring that the results generated by these platforms are reproducible and trustworthy.
Key research, such as the Research on visual analytics of multi-omics data, highlights how comprehensive platforms enable principled exploratory analysis. These publications demonstrate that when we use standardized, peer-reviewed algorithms within a unified platform, we reduce the risk of human error and “p-hacking,” leading to more robust scientific outcomes.
Stop Moving Data: Why Federated AI Scales to 100M+ Genomes
The future of biology isn’t just integrated; it’s federated. As we move toward the goal of analyzing millions of genomes across different countries and jurisdictions, we cannot keep moving data around. It’s too slow, too expensive, and too risky.
The next generation of the multi-omics data analytics platform will be driven by AI that can learn from data scattered across the globe without that data ever leaving its secure home. This is what we do at Lifebit. Our federated AI platform provides secure, real-time access to global multi-omic data, powering large-scale research for biopharma and public health agencies alike.
By using components like our Trusted Data Lakehouse (TDL) and Real-time Evidence & Analytics Layer (R.E.A.L.), we are helping researchers turn “drowning in data” into “swimming in insights.” The “Multi-Omics Marvel” isn’t just a piece of software—it’s the key to open uping the next century of human health.
Explore the next generation of federated analytics and see how we can help you turn your complex datasets into actionable biological breakthroughs.

