Drug Discovery Analytics: Turning Big Data into Big Cures

Drug Discovery Analytics: How to Cut Preclinical Costs by 30% and Stop 90% of Failures
Drug discovery analytics transforms how pharmaceutical companies and research institutions turn massive datasets into breakthrough therapies. In an era where the volume of biomedical data is doubling every few months, the ability to extract actionable insights is no longer a luxury—it is a survival requirement for the industry. This “data deluge” includes everything from high-resolution imaging and cryo-electron microscopy to longitudinal patient records and real-time sensor data from clinical trials. Here’s what modern analytics delivers:
- Predictive modeling that identifies promising drug candidates early, reducing the 90% failure rate in clinical trials by filtering out compounds with poor safety profiles before they reach human testing.
- Machine learning algorithms that analyze chemical structures, biological pathways, and patient data to predict efficacy and toxicity with unprecedented precision.
- Cost reduction of up to 30% in preclinical development time and expenses by automating routine screening and prioritizing high-probability leads.
- Real-time insights from federated data sources including genomics, Electronic Health Records (EHR), and clinical trial databases, allowing for a holistic view of patient health.
- AI-driven target identification that connects biomarkers to disease mechanisms across siloed datasets, uncovering novel therapeutic opportunities that traditional methods miss.
The pharmaceutical industry faces a brutal reality: approximately 90% of drug candidates fail in preclinical or clinical trials, often after more than a decade of development and billions of dollars invested. Most failures stem from toxicity issues or lack of efficacy that traditional methods fail to predict early enough. This “attrition crisis” has led to a significant decline in R&D productivity, where the cost of developing a new drug has skyrocketed while the number of new drug approvals remains relatively stagnant. This trend is often contrasted with the tech industry; while computing power becomes cheaper and faster, drug discovery has historically become more expensive and slower.
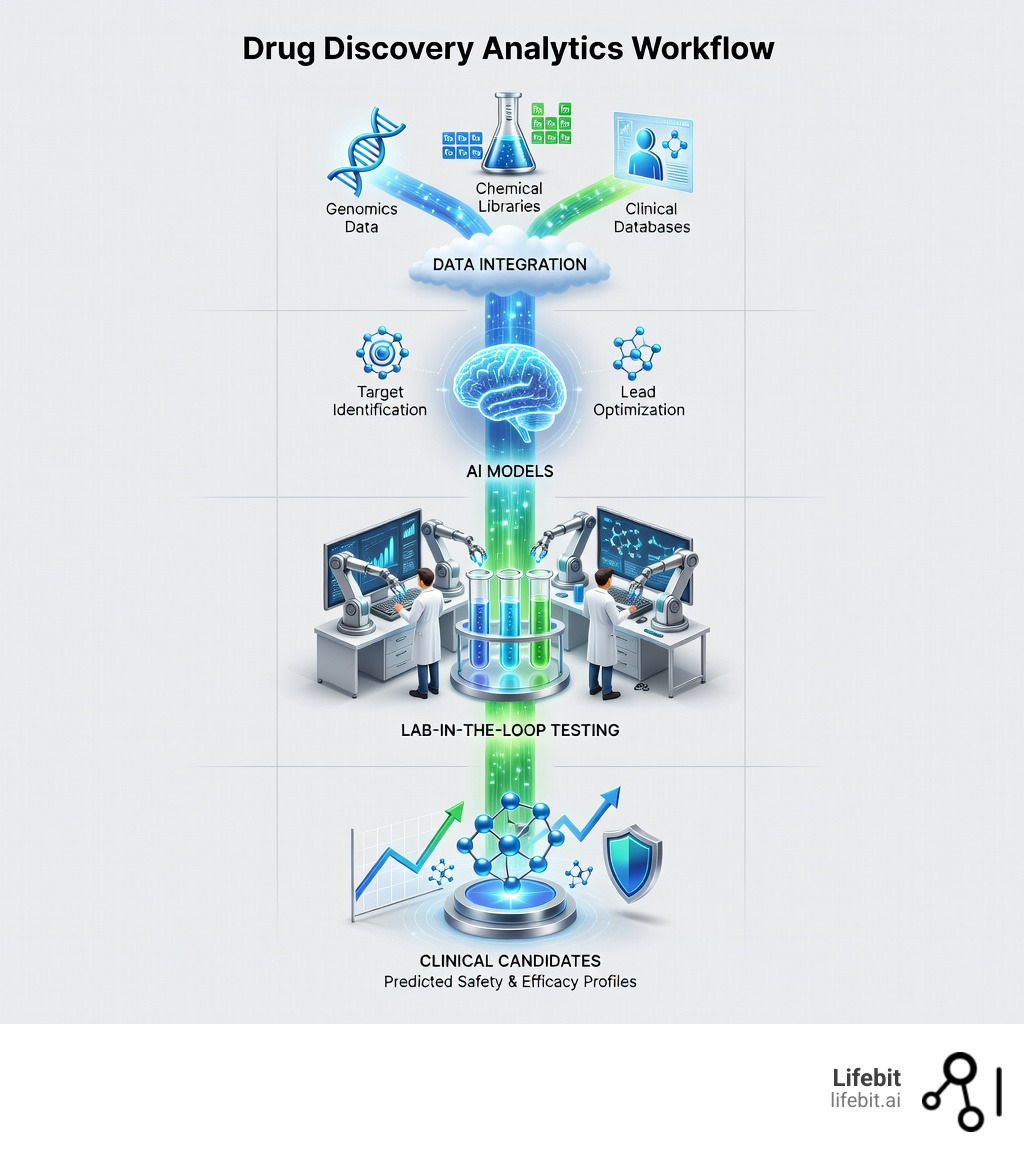
Drug discovery analytics changes this equation. By applying artificial intelligence and machine learning to vast repositories of chemical, biological, and clinical data, researchers can now predict which compounds will succeed before expensive trials begin. Organizations using AI-driven approaches have reported savings of up to 30% in time and cost during preclinical stages, while improving the probability of identifying truly effective therapies. This shift from a “fail-fast” to a “succeed-early” mentality is powered by the integration of multi-omic data—genomics, proteomics, and metabolomics—into a single analytical framework. By looking at the interplay between these different biological layers, researchers can identify “master regulators” of disease that were previously invisible.
But there’s a catch: the data challenge in drug discovery differs fundamentally from other AI success stories. While image recognition systems train on billions of labeled photos, drug discovery models often work with just hundreds of annotated compounds for critical safety endpoints. Chemical and biological data are sparse, context-dependent, and notoriously difficult to label unambiguously—a compound can be therapeutic at one dose and toxic at another, effective in one species but harmful in another. Furthermore, the “data silo” problem remains a major hurdle; valuable data is often locked within individual institutions or hidden behind proprietary walls, preventing the kind of large-scale training necessary for robust AI models.
This is where modern drug discovery analytics platforms become essential. They must integrate heterogeneous data sources—from molecular structures and protein interactions to patient genomics and real-world evidence—while maintaining compliance across regulatory boundaries. The most effective approaches combine “lab-in-the-loop” workflows, where AI predictions are iteratively validated through experiments, creating a virtuous cycle that improves model accuracy with each round. By leveraging federated learning, companies can now train models on distributed datasets without ever moving sensitive patient information, ensuring both privacy and progress. This decentralized approach allows for the creation of “global models” that benefit from the diversity of worldwide patient populations without compromising data sovereignty.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, where I’ve spent over 15 years building computational biology and AI platforms that enable secure, federated analysis of biomedical data. My work in drug discovery analytics focuses on helping pharmaceutical companies and public health institutions access and analyze siloed datasets across compliant environments, accelerating the path from variant to validated therapeutic target. We believe that the future of medicine lies in the ability to query the world’s biomedical data as if it were a single, unified database, while respecting the strict privacy requirements of every jurisdiction.
Important Drug discovery analytics terms:
- Bioinformatics data analysis
- Clinical trial data management
- Which companies offer federated AI platforms for secure analysis of biomedical data?
The $2.6 Billion Problem: Why 90% of Candidates Fail Traditional Drug Discovery
The traditional pharmaceutical R&D model is increasingly unsustainable. It currently takes more than ten years and an average investment of $2.6 billion to determine if a drug candidate is truly effective and safe for the market. During this decade-long journey, the vast majority of candidates stumble. Research shows that 90% of candidates failing drug development faces significant inefficiencies, primarily due to inefficacy or toxicity at the clinical trial phase. This is often referred to as the “Valley of Death,” the gap between initial laboratory discovery and successful clinical application where most potential cures perish. This gap is widened by the fact that many biological targets identified in academic settings are not “druggable” or fail to replicate in industrial environments.
This high attrition rate leads to astronomical financial losses and delayed patient treatments. When a drug fails in Phase III, it isn’t just a scientific setback; it represents hundreds of millions of dollars in underused R&D investment and years of lost opportunity for patients with unmet medical needs. This phenomenon is known as “Eroom’s Law” (the reverse of Moore’s Law), which suggests that drug discovery is becoming exponentially slower and more expensive over time, despite massive technological gains in other sectors. Several factors contribute to Eroom’s Law, including the “Better than the Beatles” problem, where new drugs must compete with existing, highly effective, and often off-patent therapies, raising the bar for approval and market entry.
Furthermore, “regulatory creep” has increased the complexity and volume of data required for approval. While these regulations are essential for patient safety, they demand a level of analytical rigor that traditional manual methods can no longer provide. The cost of bringing a drug to market has roughly doubled every nine years since the 1950s, creating a sustainability crisis for even the largest pharmaceutical giants. Small biotech firms are even more vulnerable, as a single late-stage failure can lead to total insolvency.
The core of this problem is a lack of R&D sustainability. Without better predictive power, we continue to waste resources on molecules that are destined to fail. Toxicity risks are often detected far too late because animal models and simple cell assays don’t always reflect the intricate complexity of human biology. For example, a compound might show no toxicity in a mouse model but cause severe liver damage in humans due to metabolic differences or specific genetic polymorphisms. To fix this, we need a fundamental shift toward data-driven decision-making that can spot these “dead ends” years earlier, using advanced analytics to simulate human biological responses more accurately than traditional methods ever could. This involves moving beyond simple correlation to understanding the causal mechanisms of drug action and adverse events.
Stop Wasting R&D: Use AI-Driven Drug Discovery Analytics to Predict Success
The integration of Artificial Intelligence (AI) and Machine Learning (ML) is finally providing the tools to break the cycle of high-cost, high-failure research. By leveraging Drug discovery analytics, we move from a trial-and-error approach to a predictive one, where data guides every step of the pipeline from target identification to clinical trial design. This transition is characterized by the use of “Digital Twins”—computational models of biological systems that can be used to test hypotheses in silico before a single pipette is touched.
The impact is already measurable and profound. Organizations using AI-driven approaches are reporting savings of up to 30% in time and cost during preclinical stages. Beyond just saving money, AI enables revenue acceleration by getting life-saving drugs to market faster, potentially adding years of patent-protected sales and, more importantly, saving lives sooner. In a competitive market, being first-to-market with a superior safety profile is the ultimate strategic advantage.
Key benefits of AI in this space include:
- Predictive Modeling: Forecasting how a molecule will behave in the human body by simulating its interaction with thousands of proteins simultaneously, identifying potential “off-target” effects that lead to side effects.
- Lead Identification: Sifting through billions of compounds in virtual libraries to find the “needle in the haystack” that possesses the perfect balance of potency, solubility, and safety.
- Resource Optimization: Focusing laboratory efforts only on the most promising candidates, thereby reducing the need for expensive and ethically sensitive animal testing and streamlining the supply chain for chemical reagents.
- ADMET Prediction: Using deep learning to predict Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties early in the design phase, which are the primary reasons for clinical failure. Modern models can now predict these properties with accuracy levels that rival or exceed traditional wet-lab assays.
Using Machine Learning for Advanced Drug Discovery Analytics
Advanced ML models are now capable of “understanding” chemistry in ways that were previously impossible. For instance, the Molecular Transformer model uses SMILES (Simplified Molecular Input Line Entry System) representations of chemical structures to predict reaction outcomes with high numerical accuracy. This allows chemists to plan complex syntheses with a much higher success rate, reducing the time spent on failed laboratory reactions and minimizing chemical waste. These models treat chemical reactions like a language translation problem, where the reactants are the “source language” and the products are the “target language.”
In the field of Artificial intelligence in drug discovery, we use Deep Learning—specifically Graph Neural Networks (GNNs)—to predict molecular properties such as solubility, permeability, and binding affinity. Unlike traditional neural networks, GNNs can process the non-Euclidean structure of molecules, treating atoms as nodes and bonds as edges. Generative chemistry takes this a step further; instead of just screening existing molecules, AI can design entirely new ones from scratch that are optimized for a specific therapeutic target. A notable success in this area was the rapid identification of potent DDR1 kinase inhibitors in just 21 days using generative AI, a process that would typically take years using traditional medicinal chemistry. This “de novo” design approach allows researchers to explore regions of chemical space that have never been synthesized before.
Bridging the Gap in Drug Discovery Analytics: In Vitro to In Vivo
One of the greatest problems in medicine is translation. A drug might look perfect in a petri dish (in vitro) but fail miserably in a living organism (in vivo). Traditional analytics often struggle here because they rely on proxy endpoints that don’t capture the full complexity of human physiology, such as the blood-brain barrier, the gut microbiome, or the intricate immune response. The human body is not a static system; it is a dynamic network of feedback loops that can compensate for or amplify a drug’s effects in unpredictable ways.
Modern Drug discovery analytics aims to close this gap by incorporating safety surveillance and pharmacovigilance data earlier in the process. By analyzing historical clinical data and side-effect targets, we can build models with higher positive predictive values (PPV). However, as research on realistic model performance suggests, we must remain healthily skeptical. Activity against a specific target is often a necessary but insufficient criterion for anticipating drug side effects. We must model the entire biological system, accounting for off-target interactions and individual genetic variability. This systems-biology approach is the cornerstone of modern analytics, ensuring that we don’t just find a molecule that binds to a protein, but a drug that works in a patient. This includes the use of “Organ-on-a-chip” data, which provides high-fidelity human biological data that can be used to refine AI predictions before human trials.
How to Solve the ‘Small Data’ Problem in Drug Discovery Analytics
Unlike fields like image or speech recognition, where data is abundant (think of the millions of images in ImageNet), drug discovery suffers from a “small data” problem for its most important endpoints. While we have millions of data points for simple chemical properties like molecular weight or logP, we may only have a handful of data points for how a specific drug affects a rare disease or a specific human sub-population. This scarcity is compounded by the high cost of generating high-quality biological labels, which often require months of specialized laboratory work.
| Feature | Image Recognition | Drug Discovery |
|---|---|---|
| Data Volume | Billions of points (ZB scale) | Hundreds to thousands for in vivo data |
| Labeling | Simple (e.g., “This is a cat”) | Complex (Dose/Context dependent) |
| Representation | Standardized pixels | Complex chemical/biological graphs |
| Space Size | High, but finite | ~10^60 plausible small molecules |
The chemical space is mind-bogglingly vast—estimated at 10^60 possible small molecules, which is more than the number of atoms in the solar system. Yet, our high-quality biological datasets are often tiny. For example, histopathology endpoints are notoriously difficult for AI because we might only have data for a few hundred compounds. Furthermore, biological labels are rarely “black and white.” A molecule’s toxicity is often dependent on the dose, the specific assay setup, or even the genetic drift of the cell lines used in the lab. This “noise” in the data can easily lead to overfitting, where a model performs perfectly on training data but fails in the real world.
To overcome this, we use multi-omic integration and Transfer Learning. By combining genomics, proteomics, and transcriptomics data, we can build a more holistic view of how a drug interacts with a biological system. Transfer learning allows us to train a model on a large, general dataset (like general chemical toxicity or protein folding) and then “fine-tune” it on a smaller, specific dataset (like toxicity in a specific lung tissue). This approach maximizes the utility of every single data point by “borrowing” knowledge from related tasks. We also employ “Few-shot learning” techniques, which are specifically designed to allow models to learn new concepts from only a handful of examples.
Another breakthrough is the use of Knowledge Graphs. These are massive networks of interconnected biological entities—genes, proteins, diseases, and drugs. By using AI to traverse these graphs, researchers can identify hidden relationships, such as a drug approved for hypertension that might also be effective for a specific type of neurodegeneration. This “drug repurposing” is a direct result of advanced analytics solving the small data problem by looking at the broader biological context. Knowledge graphs allow us to move from “black box” AI to “explainable AI,” where we can trace the path of reasoning that led to a specific prediction, which is crucial for gaining the trust of clinicians and regulators.
The Lab-in-the-Loop: How Iterative AI Creates a Virtuous Cycle for Cures
We believe the most powerful way to use AI is through the ‘lab-in-the-loop’ concept. This isn’t about replacing scientists with robots; it’s about creating a “virtuous cycle” between the computer and the bench, where each informs and improves the other in real-time. This synergy allows for “Active Learning,” where the AI system specifically requests the experiments that will most reduce its uncertainty, rather than just processing whatever data happens to be available.
In this workflow:
- AI makes a prediction: For example, it identifies 50 potential hits from a virtual library of billions based on predicted binding affinity, safety, and synthetic accessibility.
- Lab tests the prediction: Scientists synthesize and test these 50 compounds in the real world using high-throughput screening, automated microfluidics, or organ-on-a-chip technology.
- Data feeds back: The results (both successes and failures) are fed back into the AI model. Crucially, the “failures” are often more informative than the successes, as they teach the model where its boundaries lie and help it map the “negative space” of the chemical landscape.
- Model improves: The AI learns from the real-world results, adjusts its weights, and makes even better, more refined predictions in the next round, often suggesting modifications to the chemical structure to improve potency or reduce toxicity.
This active learning approach requires close collaboration between technology companies and pharmaceutical researchers. It also demands a robust strategy development phase, where organizations prioritize which biological questions are most likely to benefit from AI intervention. At Lifebit, we facilitate this through our Trusted Research Environment (TRE), allowing secure, real-time insights across hybrid data ecosystems. A TRE provides a secure “walled garden” where researchers can bring their tools to the data, rather than moving the data to the tools. This is essential for maintaining data sovereignty and complying with international data protection laws like GDPR.
By keeping the data where it lives and bringing the analysis to the data, we enable a global, collaborative lab-in-the-loop that can tackle the world’s most challenging diseases. This architecture also supports “Federated Learning,” where multiple institutions can collaborate on a single model without ever sharing their proprietary raw data. This breaks down the competitive silos that have historically slowed down drug discovery, allowing the industry to move forward as a collective while still protecting individual intellectual property. The result is a faster, more efficient, and more secure path to the clinic.
Frequently Asked Questions about Drug Discovery Analytics
What are the primary challenges in drug discovery analytics?
The three biggest problems are data scarcity (especially for in vivo safety data), biological complexity (the unpredictable nature of human systems), and translation failure (the difficulty of moving from lab findings to clinical success). Additionally, data interoperability—ensuring that data from different labs, machines, and historical eras can be analyzed together—remains a significant technical hurdle. Standardizing metadata and ensuring data quality are constant battles for data scientists in this field.
How does AI improve pharmaceutical R&D sustainability?
AI drives sustainability by significantly reducing cost and time. By avoiding late-stage failures, companies minimize the waste of resources, capital, and animal lives. AI-driven screening can reduce the number of physical experiments needed by orders of magnitude, allowing researchers to explore a much wider range of therapeutic possibilities with a smaller environmental and financial footprint. It also allows for the repurposing of existing drugs, which is the most sustainable way to find new treatments.
What is the ‘lab-in-the-loop’ concept?
It is an iterative AI framework where laboratory experiments and computational models are inextricably linked. The lab provides the “ground truth” data that trains the AI, while the AI provides the “intelligent guidance” that tells the lab which experiments are worth running. This prevents the “garbage in, garbage out” problem that plagues many static AI models and ensures that every dollar spent in the lab generates maximum scientific value.
Can AI predict drug-drug interactions?
Yes, modern analytics can predict how different drugs will interact within the body by modeling their metabolic pathways and competition for specific enzymes (like the Cytochrome P450 family). This is crucial for elderly patients or those with chronic conditions who are often taking multiple medications simultaneously (polypharmacy), reducing the risk of adverse drug events before they occur in the population.
Is data privacy a concern in drug discovery analytics?
Absolutely. Patient genomic and health data are highly sensitive and protected by strict laws. This is why federated learning and Trusted Research Environments (TREs) are so important. They allow for robust analysis and model training without ever exposing or moving the underlying raw data, ensuring full compliance with regulations like GDPR and HIPAA while still allowing for global scientific collaboration.
What is Explainable AI (XAI) in drug discovery?
Explainable AI refers to techniques that allow humans to understand why an AI model made a specific prediction. In drug discovery, this might mean identifying the specific part of a molecule that the AI thinks is toxic or the specific gene pathway it believes is being targeted. XAI is critical for regulatory approval, as agencies like the FDA often require a mechanistic understanding of how a drug works rather than just a statistical prediction of success.
The Future of Medicine: Secure, Federated Drug Discovery Analytics
The next decade will be the most transformative in the history of medicine. As we move toward 2030, the impact of Drug discovery analytics on human health will be profound. We are moving away from a world of “blockbuster” drugs designed for the average person and toward a future of precision medicine designed for the individual. In this future, a patient’s own genetic data, lifestyle factors, and even their microbiome profile will be used to select the drug and dose that will be most effective for them with the fewest side effects. This is the promise of “Personalized Medicine,” and it is entirely dependent on advanced analytics.
At Lifebit, we are proud to be at the forefront of this revolution. Our federated AI platform and Trusted Research Environment provide the secure infrastructure needed for global collaboration. By enabling researchers to analyze multi-omic data where it lives, we ensure that privacy and compliance never stand in the way of a cure. We are also exploring the potential of Quantum Computing to simulate molecular interactions at the atomic level with perfect precision. While still in its early stages, quantum computing could eventually solve the “Schrödinger equation” for large molecules, providing even more accurate data for our AI models and potentially eliminating the need for many early-stage physical experiments.
Furthermore, the rise of “Digital Twins” for patients will allow doctors to simulate the effects of a drug on a virtual version of a patient before prescribing it. This will virtually eliminate the trial-and-error process that currently defines many treatment regimens, particularly in oncology and psychiatry. The ethical use of AI will also be a major focus, ensuring that the benefits of these technologies are distributed equitably across different global populations and that the models themselves are free from bias.
The road to a new drug is long and fraught with risk, but with the right analytics, we can turn the “big data” of today into the “big cures” of tomorrow. The goal is not just to make drug discovery faster or cheaper, but to make it more human—ensuring that every patient has access to the treatments they need, when they need them. We are entering an era where the only limit to medical progress is our ability to analyze the data we already have.

