The Ultimate Guide to Trusted Research Environments

Access 58M Patient Records Without Risk: Use a Trusted Research Environment TRE
In the current landscape of biomedical innovation, we are witnessing a profound “Data Paradox.” On one hand, the volume of health data is growing exponentially, with petabytes of genomic sequences and clinical records being generated annually. On the other hand, accessing this data has never been more difficult due to the necessary tightening of privacy laws and the increasing threat of cyberattacks. A trusted research environment TRE is the definitive solution to this paradox. It is a highly secure digital platform that enables approved researchers to remotely access and analyze sensitive biomedical data—such as genomic sequences, electronic health records (EHR), and clinical trial results—without compromising patient privacy or regulatory compliance.
Historically, data sharing involved the physical or digital transfer of datasets from a custodian to a researcher. This “data download” model is inherently risky; once the data leaves the custodian’s server, control is lost. A TRE flips this model on its head. Instead of moving the data to the researcher, the TRE brings the researcher to the data. This shift from “Data as a Product” to “Data as a Service” ensures that the data remains within a secure, audited perimeter at all times.
Key features of a TRE include:
- Secure remote access – Researchers use encrypted connections to analyze data inside the environment; the raw data never leaves the secure perimeter, only aggregated results can be exported after review.
- De-identified and Pseudonymized datasets – All direct identifiers, such as names, social security numbers, and exact addresses, are removed. Advanced techniques like k-anonymity are often applied to ensure individuals cannot be re-identified through combination with other datasets.
- Controlled permissions and Role-Based Access Control (RBAC) – Access is not binary. Only trained, approved researchers with specific ethical clearance can access the specific subsets of data required for their approved project.
- Exhaustive Audited activity – Every action, from a simple file view to a complex SQL query or a machine learning training run, is logged. This creates a transparent, immutable audit trail for data custodians and regulators.
- Multi-modal support and Data Integration – Modern TREs are not just for spreadsheets. They integrate high-depth genomics (WGS/WES), medical imaging (DICOM), longitudinal EHRs, and real-world evidence (RWE) in a single, high-performance workspace.
TREs are also frequently referred to as Secure Data Environments (SDEs) or Data Safe Havens. They solve the fundamental challenge of modern medicine: how to unlock the immense scientific value of sensitive health data at scale while meeting the strict requirements of the General Data Protection Regulation (GDPR), the Health Insurance Portability and Accountability Act (HIPAA), and the emerging European Health Data Space (EHDS).
Why TREs matter now:
The complexity of health data has outpaced traditional infrastructure. UK Biobank now holds whole-genome sequencing data from 500,000 individuals, a dataset so large that downloading it is practically impossible for most academic institutions. OpenSAFELY provides secure access to NHS records for over 58 million people in England, allowing for unprecedented population-level analysis. Genomics England has enabled more than 2,000 researchers to analyze data from the 100,000 Genomes Project, leading to new diagnoses for rare disease patients.
Without a robust trusted research environment TRE, researchers face significant hurdles:
- Legal and Administrative Bottlenecks: Negotiating Data Transfer Agreements (DTAs) can take years.
- Infrastructure Sprawl: Deploying separate, non-interoperable tools for every new dataset leads to wasted resources.
- Security Vulnerabilities: Moving sensitive data increases the surface area for potential breaches.
- Collaboration Barriers: Siloed data prevents the cross-institutional studies necessary for identifying rare genetic variants.
As CEO and Co-Founder of Lifebit, I’ve spent over 15 years building the platforms that power secure, federated analysis of global biomedical data. My work with public sector institutions and global pharmaceutical organizations has shown that the trusted research environment TRE is no longer an optional luxury—it is the essential infrastructure for the future of precision medicine. This guide will walk you through the technical frameworks, economic benefits, and real-world applications of TREs, providing a roadmap for ethical, scalable health data research.
Stop Data Downloads: Use a Trusted Research Environment TRE Instead
At its core, a secure computing environment known as a TRE is designed to function like a high-security digital vault. The traditional model of data sharing is akin to a library that lets you take rare, fragile manuscripts home; the risk of loss or damage is high. In contrast, a TRE is like a reading room where you can study the manuscripts under supervision, take notes on the insights you find, but the original documents never leave the room. This “data stays put” philosophy is the defining characteristic of a trusted research environment TRE.
The life sciences industry is currently facing a crisis of “data gravity.” As datasets grow into the petabyte scale, the cost and time required to move them become prohibitive. Furthermore, the legal landscape has shifted. Global regulations like GDPR in Europe and HIPAA in the USA mandate that patient data must be protected with the highest level of rigor, often requiring that data remains within specific geographic borders (data sovereignty). A TRE allows organizations to comply with these laws by providing a localized, secure access point that satisfies both legal and technical requirements.
Beyond security, TREs address the issue of tool sprawl. In many research settings, scientists are forced to use a fragmented array of cloud providers, local servers, and proprietary software, each with its own security protocols and data formats. This fragmentation slows down discovery and increases the risk of human error. A modern TRE consolidates these resources into a single, unified workspace, providing the secure data sharing framework necessary for international collaboration without the risk of data leakage.

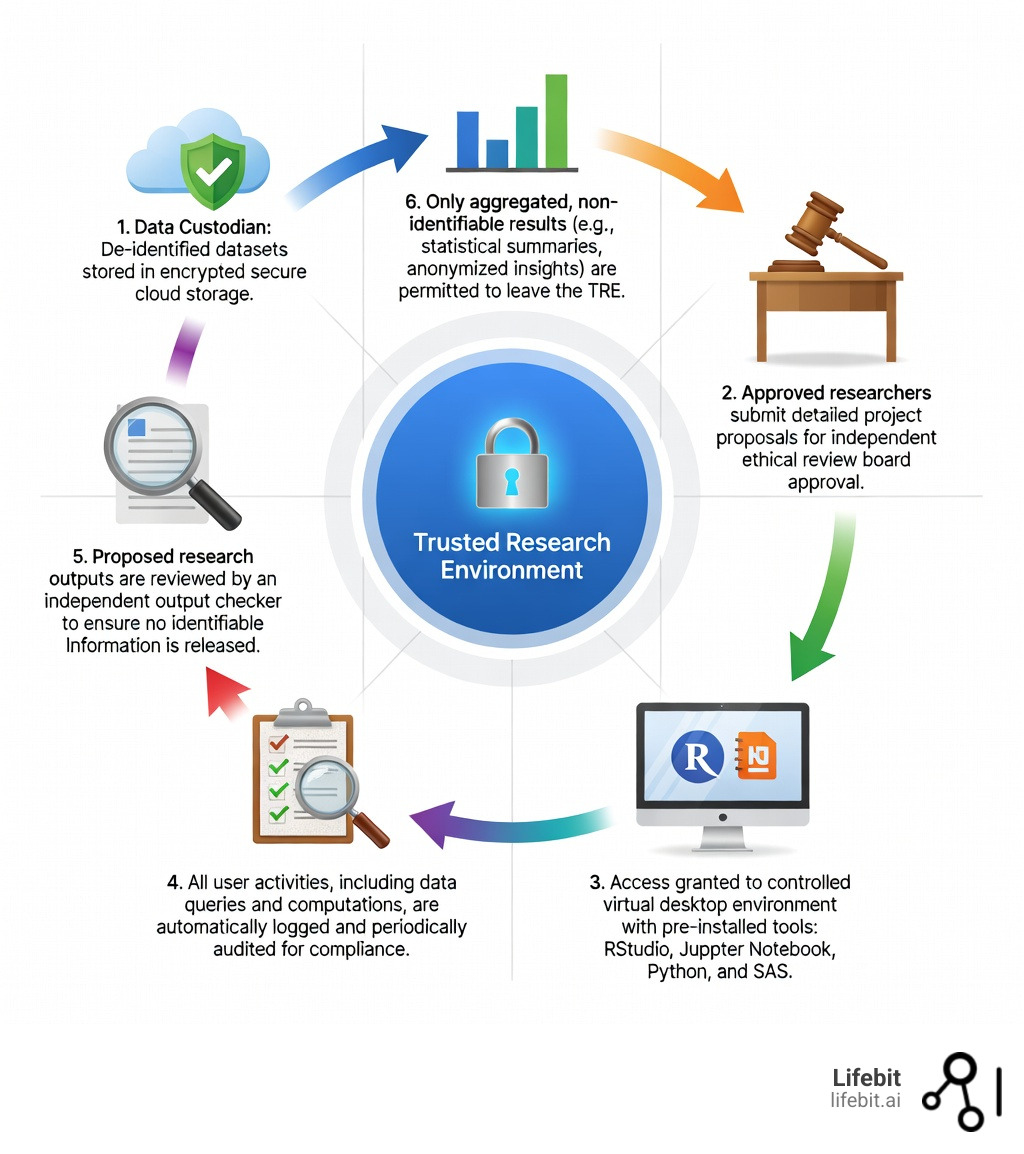
The Five Safes Framework: The Gold Standard for a Trusted Research Environment TRE
To ensure that “trust” is a measurable technical reality rather than just a marketing term, the industry relies on the Five Safes Framework. Originally developed by the UK Office for National Statistics, this framework is the foundational architecture we use at Lifebit to build a trusted research environment TRE.
-
Safe People: This pillar ensures that only verified individuals can enter the environment. This involves multi-factor authentication (MFA), identity federation (e.g., using institutional logins), and mandatory training on data privacy and ethics. Researchers must often sign legally binding data access agreements that hold them personally accountable for their conduct within the TRE.
-
Safe Projects: Access is not granted for general “browsing.” Every research project must have a clearly defined, ethical purpose that has been vetted by an independent Data Access Committee (DAC) or Institutional Review Board (IRB). The project must demonstrate that the use of sensitive data is necessary and that the potential public health benefits outweigh the privacy risks.
-
Safe Settings: The environment itself is a “walled garden.” It is technically isolated from the public internet. Inbound and outbound traffic is strictly controlled through “airlocks.” Researchers cannot simply copy-paste data out of the environment or upload unauthorized software. The TRE provides a pre-configured suite of tools (like RStudio, Jupyter, and Nextflow) within a secure virtual desktop infrastructure (VDI).
-
Safe Data: Data is treated to minimize the risk of disclosure before it is even made available. This involves de-identification (removing direct identifiers) and pseudonymization (replacing identifiers with unique codes). In more advanced setups, researchers might work with “synthetic data” for code development before running their final analysis on the real, sensitive datasets. Techniques like differential privacy may also be used to add mathematical “noise” to the data, ensuring that the presence of any single individual cannot be confirmed.
-
Safe Outputs: This is the final line of defense. Before any results, charts, or tables can be exported from the TRE, they must undergo an output check. This process, often a mix of automated scripts and manual review by data custodians, ensures that the results are truly aggregate and do not inadvertently reveal information about small groups of individuals (e.g., a table showing a rare disease result for a specific zip code where only one patient lives).
By strictly adhering to these five pillars, a TRE provides a comprehensive risk mitigation strategy that ensures all research remains in ethical alignment with patient consent and public trust.
Cut Research Costs by 70% with a Trusted Research Environment TRE
The economic argument for a trusted research environment TRE is as compelling as the security argument. The shift from “data downloads” to “bringing researchers to the data” offers transformative advantages in terms of operational efficiency and capital expenditure. In the legacy model, a pharmaceutical company might spend six to twelve months negotiating a data transfer agreement, only to find that the data format is incompatible with their internal systems or that the compute power required to analyze it is unavailable.
1. Faster Time to Insights and Scalable Compute:
By leveraging cloud providers like AWS or Microsoft Azure, TREs provide massive, on-demand scalable compute. Researchers can spin up clusters of hundreds of CPUs or GPUs to run complex genomic pipelines or deep learning models in hours rather than weeks. This elasticity means organizations only pay for the compute they use, eliminating the need for expensive, underutilized on-premise hardware.
2. Improved Reproducibility and Traceability:
Scientific integrity relies on the ability to reproduce results. Inside a TRE, every version of a dataset, every script run, and every software container used is logged and version-controlled. This creates a perfect audit trail and a “provenance map” for every discovery. This level of traceability is a critical requirement for regulatory submissions to bodies like the FDA or EMA, where the journey from raw data to clinical insight must be fully transparent.
3. Reduced Infrastructure and Maintenance Costs:
Maintaining a secure data center that meets modern compliance standards is an enormous financial burden. By adopting a trusted research environment TRE, organizations can offload the heavy lifting of security patching, hardware maintenance, and network management to the platform provider. This allows research teams to focus on science rather than IT infrastructure.
4. Data Harmonization and Interoperability:
One of the greatest hidden costs in research is “data cleaning.” TREs often include built-in tools to standardize data into common formats like the OMOP Common Data Model (CDM) or HL7 FHIR. This means a researcher can seamlessly combine a genomic dataset from a UK cohort with clinical data from a US hospital, creating a “harmonized” view that is significantly more powerful than the individual parts. This interoperability is essential for the large-scale meta-analyses required to identify subtle genetic drivers of disease.
Accelerating Discovery with a Trusted Research Environment TRE
In the competitive world of drug discovery and biomarker identification, speed is the primary differentiator. A trusted research environment TRE allows translational medicine teams to analyze multi-modal data—such as RNA-seq, digital pathology images, and longitudinal EHR data—in a single collaborative space.
For population health and genomic medicine, TREs are the only viable way to study rare diseases. Because rare disease patient populations are small and geographically dispersed, researchers must connect cohorts from multiple countries to achieve statistical significance. A federated TRE allows a researcher to run an analysis across ten different national databases simultaneously without the data ever crossing a border. This is the key to unlocking real-world evidence (RWE) and improving patient outcomes at a global scale, ensuring that no patient is left behind simply because their data is stored in a different jurisdiction.
4 Medical Breakthroughs Powered by a Trusted Research Environment TRE
TREs are no longer a theoretical concept; they are the operational engine behind some of the most significant medical breakthroughs of the last decade. By providing a secure bridge between data custodians and the global scientific community, these environments have fundamentally changed the pace of discovery.
-
UK Biobank: This is perhaps the most famous example of a TRE in action. Containing de-identified genomic, imaging, and lifestyle data from 500,000 participants, it is a goldmine for chronic disease research. To protect this resource, UK Biobank launched its Research Analysis Platform (RAP), a cloud-based TRE that allows researchers to bring their code to the data. This has led to thousands of peer-reviewed publications on everything from the genetic basis of heart disease to the impact of COVID-19 on brain structure.
-
Yale University Open Data Access (YODA) Project: The YODA Project has pioneered the movement for clinical trial transparency. By providing a secure TRE for the analysis of clinical trial data from 491 pharmaceutical trials (including those from Johnson & Johnson), it has enabled independent scientists to validate drug safety and efficacy. This model of open science, conducted within a “Safe Setting,” has set a new standard for the pharmaceutical industry.
-
Genomics England Research Environment: As part of the 100,000 Genomes Project, Genomics England created a TRE that currently supports over 2,000 researchers. This platform has been instrumental in identifying new genetic variants responsible for rare diseases and tailoring cancer treatments to the specific genetic profile of a patient’s tumor. It demonstrates how a TRE can serve as a direct link between research and clinical care.
-
Rare Disease Cures Accelerator (RDCA-DAP): Funded by the US FDA and managed by the Critical Path Institute, this platform uses a TRE to aggregate data from disparate rare disease trials and registries. By providing a unified environment for analysis, it helps researchers understand the natural history of rare diseases, which is a prerequisite for designing successful clinical trials for new treatments.
At Lifebit, our Lifebit Trusted Research Environment powers similar large-scale initiatives. A standout example is the OpenSAFELY platform. During the COVID-19 pandemic, OpenSAFELY used a TRE approach to analyze the full, pseudonymized primary care records of 58 million people in England. This allowed researchers to provide near real-time insights into COVID-19 risk factors and vaccine effectiveness without the data ever leaving the NHS secure servers. Other notable examples include EPIC-Norfolk, which provides lifestyle and health data from over 30,000 participants through its secure research computing platform, enabling long-term studies on aging and nutrition.
Best Practices for Implementing Secure Research Infrastructure
Building a TRE is a complex multi-disciplinary undertaking. Based on our extensive experience and the Lifebit TRE Guide 2026, here are the essential best practices for a successful implementation:
- Prioritize Cloud-Native Architecture: Avoid the trap of “lift and shift.” Use cloud-native technologies like Kubernetes for orchestration and Docker for containerization. This ensures your environment is scalable, resilient, and can be updated without downtime.
- Embed FAIR Principles: Data is only useful if it is Findable, Accessible, Interoperable, and Reusable. A TRE should include a robust metadata catalog that allows researchers to understand the context of the data they are analyzing. This is a prerequisite for AI/ML readiness.
- Support Multi-Modality from Day One: Modern research is multi-dimensional. Your TRE must be able to handle more than just tabular data; it needs optimized storage and compute for high-resolution pathology images, complex genomic VCF files, and unstructured clinical notes.
- Implement Granular, Attribute-Based Access Control (ABAC): Move beyond simple “on/off” access. Use fine-grained permissions that restrict users to specific rows, columns, or files based on their project’s specific ethical approval.
- Integrate Standard, Open-Source Tools: Researchers are most productive when using tools they already know. Ensure your TRE supports RStudio, Jupyter Notebooks, and workflow languages like Nextflow and WDL.
- Plan for GPU and AI Acceleration: The future of research is AI-driven. Your infrastructure must be capable of spinning up high-performance GPUs (like NVIDIA A100s) on demand to support deep learning and large language model (LLM) training.
The AI Shortcut: Why a Federated Trusted Research Environment TRE Wins
The next frontier for TREs is the seamless integration of Artificial Intelligence (AI) and Machine Learning (ML). As noted by McKinsey, AI represents a “once-in-a-century opportunity” for pharma companies to reshape how we design proteins, identify drug targets, and stratify patients for clinical trials. However, the bottleneck for AI has always been data access. Training a robust AI model requires vast, diverse datasets that no single institution possesses.
This is where the federated trusted research environment becomes a game-changer. In a traditional centralized model, you would have to move data from multiple hospitals or countries into one central location to train a model—a process that is often legally and technically impossible. In a federated model, we move the model to the data.
How Federated Learning Works in a TRE:
- A central “orchestrator” sends a base AI model to multiple secure TREs (the “worker nodes”) located at different data sites.
- The model is trained locally on the sensitive data within each TRE.
- Only the model “weights” (the mathematical updates, not the raw data) are sent back to the orchestrator.
- The orchestrator aggregates these updates to create a smarter, more accurate global model.
This approach allows for “collaborative AI” where a model can learn from millions of patients across the globe without a single byte of sensitive raw data ever being exposed or crossing a border. It is the ultimate expression of the “Safe Data” and “Safe Settings” pillars of the Five Safes framework.
Future-proof TREs will also focus on several emerging trends:
- EHDS Compliance: Preparing for the European Health Data Space, which will mandate standardized ways for researchers to access health data across all EU member states.
- TileDB and Array-Based Storage: Moving away from legacy file formats to array-based storage like TileDB, which allows for lightning-fast queries across massive genomic “variant warehouses.”
- Real-Time Collaborative Coding: Enabling teams of bioinformaticians across different continents to work on the same code, share live dashboards, and troubleshoot pipelines in a synchronized, secure workspace, drastically reducing the time from hypothesis to discovery.
Trusted Research Environment TRE: Stop Compliance Risks and Tool Sprawl
How do TREs ensure GDPR, HIPAA, and EHDS compliance?
TREs achieve compliance through a “defense-in-depth” strategy, where multiple layers of security work together to protect the data. According to our Trusted Research Environments Explained guide, this includes:
- Advanced De-identification: Beyond just removing names, TREs use techniques like date-shifting and generalization to ensure that even sophisticated “linkage attacks” cannot re-identify individuals.
- Digital Airlock Systems: These are strict digital barriers. Any file a researcher wants to import (like a custom script) or export (like a summary table) must pass through the airlock, where it is scanned for malware and reviewed for disclosure risk.
- End-to-End Encryption: Data is encrypted using industry-standard protocols (like AES-256) while it is stored (“at rest”) and while it is being processed or moved between secure nodes (“in transit”).
- Immutable Audit Logs: Every single command entered into a terminal, every file opened, and every API call is recorded in a tamper-proof log. This provides data custodians with the absolute certainty they need to satisfy regulators and internal auditors.
What are the main limitations of current TRE systems?
While the technology has advanced rapidly, there are still challenges that the industry, including the UK Health Data Research Alliance, is working to solve:
- Semantic Interoperability: Even if two TREs use the same security protocols, they may use different coding systems for clinical data (e.g., ICD-10 vs. SNOMED-CT). This makes cross-platform analysis difficult without significant manual data mapping.
- The “Human Bottleneck” in Output Checking: In many legacy TREs, the final check of research results is a manual process performed by a human data manager. This can create a backlog, delaying the publication of important findings. The industry is moving toward “automated disclosure control” to speed this up.
- User Experience (UX) Friction: Some early TREs were notoriously difficult to use, offering clunky interfaces and limited software libraries. Modern TREs must prioritize the researcher’s experience, providing a seamless, high-performance environment that feels like their local machine.
- Cost of Egress and Compute: While cloud-native TREs are more efficient, the sheer scale of modern genomic analysis means that compute costs can still be significant. Optimizing pipelines for cost-efficiency is a major area of ongoing research.
What role do cloud-native tools like TileDB play in modern TREs?
Tools like TileDB are revolutionizing the backend of the trusted research environment TRE. Traditional genomic file formats (like VCF or BAM) were designed for single-sample analysis and are incredibly slow when you try to query them across a population of 500,000 people. TileDB uses a multi-dimensional array format that allows for:
- Sub-Second Random Access: A researcher can find a specific genetic variant across an entire population-scale dataset in seconds, rather than hours.
- Unified Metadata Management: It allows complex clinical information (like age, disease status, and medication history) to be stored alongside the genomic data, enabling much faster multi-modal queries.
- Extreme Scalability: As datasets grow from thousands to millions of genomes, array-based storage scales linearly, ensuring that the TRE remains performant even under massive loads.
Organizations like BeginNGS are already using these technologies to manage rare disease data for newborn sequencing, ensuring that the infrastructure can keep up with the life-saving speed of clinical decision-making.
Unlock Global Health Data with a Trusted Research Environment TRE
The era of siloed, inaccessible, and risky health data sharing is coming to an end. At Lifebit, we believe that the trusted research environment TRE is the fundamental key to unlocking a new age of medical discovery. By moving away from the dangerous “data download” model and embracing a “data stays put” philosophy, we can finally bridge the gap between the necessity of data security and the urgency of scientific innovation.
A modern TRE is more than just a secure server; it is a collaborative ecosystem. It provides researchers with the high-performance tools they need, gives data custodians the absolute control they require, and ensures that patients’ privacy is protected by the most advanced technical safeguards available. Whether you are a pharmaceutical company looking to accelerate your drug development pipeline, a research institute aiming to lead in genomic medicine, or a government agency tasked with protecting citizen data, the TRE is your path forward.
By embracing the Five Safes, leveraging cloud-native technologies like Kubernetes and TileDB, and preparing for a federated, AI-driven future, we can turn the world’s vast repositories of health data into life-saving knowledge—securely, ethically, and responsibly. The technology is here, the regulatory frameworks are maturing, and the potential for human health is limitless.
To learn more about how we can help you build, manage, or access a state-of-the-art research environment that meets the highest global standards, explore the Lifebit Platform today and join the revolution in secure biomedical research.

