Why You Should Never Skip Drug Target Validation

Drug Target Validation: Stop the 50% Failure Rate and Save Billions
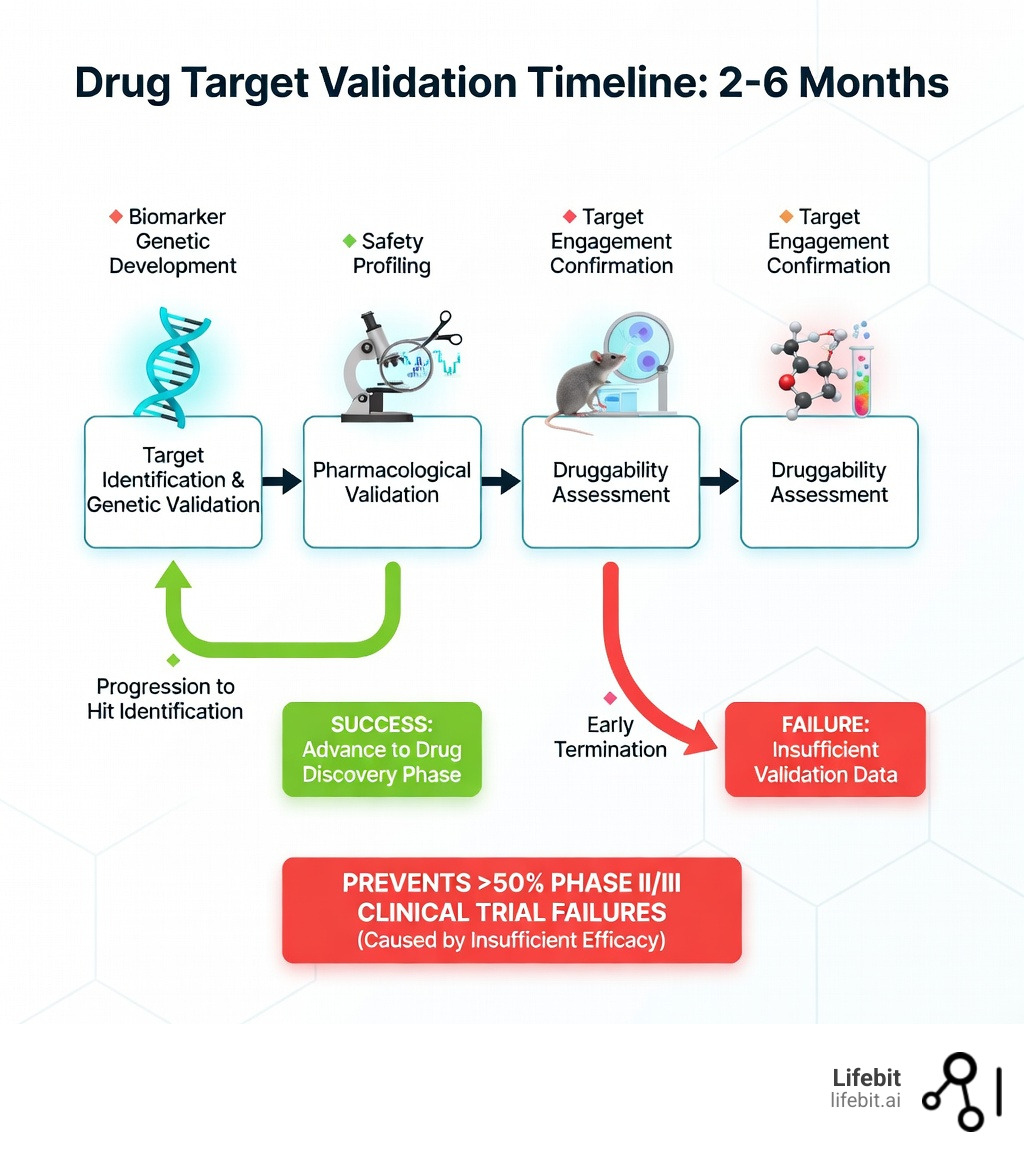
Drug target validation is the process of confirming that a specific biological molecule—like a protein, enzyme, or gene—actually causes disease and can be safely modulated by a drug. In the high-stakes world of pharmaceutical R&D, this is the most critical gatekeeping step. It transforms a biological hypothesis into a viable therapeutic strategy. To properly validate a therapeutic target, researchers must establish a multi-dimensional “weight of evidence” that includes:
- Biological confirmation: Demonstrating that the target plays a direct, functional role in the disease’s pathophysiology. This involves showing that the target’s activity is altered in diseased states and that its modulation reverses or halts disease progression.
- Genetic evidence: Utilizing human genetics to link the target to patient outcomes. This includes Genome-Wide Association Studies (GWAS), Mendelian randomization, and knockout studies that provide a causal link between genetic variants and disease risk.
- Pharmacological proof: Showing that chemical modulation of the target improves disease markers in both in vitro (cell-based) and in vivo (animal) models. This requires high-quality chemical probes that are selective for the target.
- Safety assessment: Evaluating the potential for “on-target” toxicity. If a target is essential for normal physiological function in healthy tissues, modulating it may cause unacceptable side effects.
- Druggability analysis: Confirming the target possesses structural features, such as well-defined binding pockets, that allow it to interact with small molecules, antibodies, or other therapeutic modalities.
This process typically takes 2-6 months but can prevent billions in wasted investment. The pharmaceutical industry is currently grappling with a productivity crisis; more than half of drugs fail in Phase II and III clinical trials due to insufficient efficacy—a direct result of inadequate target validation. In fact, attrition in Phase II alone is approximately 66%, driven by failures in efficacy, safety, and pharmacokinetics. When pharmaceutical companies like Bayer examined their internal validation studies, they found that only 20-25% of published literature data on potential drug targets were reproducible.
The financial stakes are enormous. Modern drug discovery costs upwards of $800 million to $2.6 billion per approved drug and takes at least 12 years. Most of that cost is back-loaded into late-stage clinical trials—trials that could be avoided if targets were properly validated early. As one workshop summary from the Institute of Medicine noted, establishing pharmacologically relevant exposure levels and target engagement are the two most critical steps in validation. Without them, even the most promising compounds are destined to fail.
I’m Maria Chatzou Dunford, CEO and Co-founder of Lifebit, with over 15 years of experience in computational biology, AI, and genomics—fields that now power modern drug target validation through federated data analysis and multi-omic integration. Throughout my career building tools for precision medicine, I’ve seen how robust target validation separates successful drug programs from costly failures. By integrating real-world evidence and advanced analytics, we can move beyond the “reproducibility crisis” and build a more efficient pipeline for the next generation of cures.
Drug target validation word roundup:
Cut Phase II Failures: How Target Validation De-Risks Your Pipeline
We have all heard the horror stories of the “Valley of Death” in drug development. This is the precarious gap between basic laboratory research and clinical application. A researcher finds a promising protein in a lab, a startup raises millions, and five years later, the Phase II trial results come back as a flatline. The drug was safe, but it simply didn’t work. Why? Because the original biological hypothesis was never properly validated in a human-relevant context. The target was associated with the disease, but it wasn’t the driver of the disease.
The process of target validation is the rigorous scientific “stress test” that follows target identification. While identification asks, “Which molecule is associated with this disease?”, validation asks, “If we hit this molecule, will the patient actually get better?” This distinction is vital. Many targets identified through differential gene expression analysis are merely “bystanders”—molecules whose levels change as a result of the disease, rather than molecules that cause the disease.
By performing drug target validation early, we move from correlation to causation. This isn’t just about good science; it’s about survival in a competitive market. Reducing Phase II failures is the single most effective way to lower the cost of developing new molecular entities. This economic reality is often referred to as “Eroom’s Law”—the observation that drug discovery is becoming slower and more expensive over time, despite improvements in technology. Robust validation is the only way to reverse this trend. When we validate a target, we are essentially de-risking the entire downstream pipeline, ensuring that the clinical trial safety profile and therapeutic modulation are backed by robust, reproducible evidence. This involves using human-derived models, such as organoids or primary patient cells, to ensure that the biology observed in the lab translates to the complex environment of the human body.
4 Traits of a Successful Drug Target: Avoid the ‘Undruggable’ Trap
Not all biological molecules are created equal. In the early days of drug discovery, researchers focused on the “low-hanging fruit”—targets that were easily accessible and well-understood. Today, we must be more strategic. Some molecules are “druggable,” meaning they have accessible binding sites where a small molecule or biologic can latch on and do its job. Others are “undruggable”—at least with our current toolkit. So, what makes an optimal target?
First, an ideal target must have a pivotal role in the pathophysiology of the disease. If the target is just a bystander, modulating it won’t change the disease’s course. We also look for tissue specificity. If a target is expressed everywhere in the body, hitting it might cure the disease in the lungs but cause a disaster in the liver. This is why G protein-coupled receptors (GPCRs) and kinases have historically been the “gold standard” targets—they have clear binding pockets and well-understood signaling roles. However, even within these classes, validation is required to ensure that the specific isoform being targeted is the one responsible for the disease state.
Modern drug target validation now explores RNA targets and protein-protein interactions (PPIs), which were once considered impossible to hit. The blueprint for success includes:
- Druggability: A 3D structure or binding pocket that can be targeted. This often involves Lipinski’s Rule of Five for small molecules, ensuring the compound has the right physical properties to be absorbed and reach its target.
- Genetic Evidence: Naturally occurring human mutations that mimic the drug’s intended effect. For example, individuals with natural “loss-of-function” mutations in the PCSK9 gene have lower cholesterol and a lower risk of heart disease, which validated PCSK9 as a prime target for cholesterol-lowering drugs.
- Safety Profile: Low risk of “on-target” toxicity in healthy tissues. This requires a deep dive into the target’s expression profile across the entire human proteome.
- Commercial Viability: A clear path to fulfilling an unmet medical need. A target might be scientifically sound, but if the patient population is too small or the existing treatments are already highly effective and cheap, the target may not be commercially viable.
CRISPR to CETSA: Validate Drug Targets in 2-6 Months

The days of relying solely on basic cell cultures are over. Today, we use a sophisticated mix of wet-lab and dry-lab techniques to build a “weight of evidence” for a target. This often begins with portfolio assessment tools that help us rank targets based on their likelihood of success, considering factors like genetic association scores and druggability indices.
One of the most powerful tools in our arsenal is the use of biomarkers. By identifying a measurable signal—like a specific protein level in the blood or a change in gene expression—that changes when the target is modulated, we can prove target engagement long before we see a clinical cure. In vitro techniques allow us to test these interactions in disease-relevant cell lines, including 3D models and iPSCs (induced pluripotent stem cells) that better mimic human biology than traditional immortalized cell lines.
In Vitro and In Vivo Drug Target Validation Techniques: Testing in the Real World
To truly validate a target, we have to see how it behaves in a living system. In vivo validation typically involves mouse models, which remain a highly reliable system for confirming potential targets, especially in oncology. Tumor cell line xenograft models allow us to see how a drug interacts with human-like tumors in a complex biological environment. More advanced models, such as Patient-Derived Xenografts (PDX), involve transplanting actual patient tumor tissue into mice, providing a more accurate representation of human disease heterogeneity.
We also use advanced cell-based assays like CETSA (Cellular Thermal Shift Assay). CETSA is a game-changer because it measures drug-protein binding directly in the cell by monitoring changes in the protein’s thermal stability. When a drug binds to a protein, it typically stabilizes the protein’s structure, making it more resistant to heat-induced denaturation. By measuring this “thermal shift,” researchers get undeniable proof of target engagement—one of the biggest hurdles in early discovery. Other essential methods include:
- CRISPR-Cas9: This gene-editing technology allows for precise “knockout” of specific genes. If removing the gene stops the disease phenotype in a cell model, it provides strong evidence that the gene’s protein product is a valid target. CRISPR can also be used for “knock-in” experiments to introduce specific disease-associated mutations.
- RNA Interference (RNAi): Using siRNA or shRNA to temporarily silence gene expression. This allows researchers to observe the functional changes that occur when target levels are reduced, mimicking the effect of an inhibitory drug.
- Pharmacological Probes: Using high-quality, highly selective “tool molecules” to see if chemical inhibition produces the same result as genetic silencing. If a chemical inhibitor and a genetic knockout produce the same biological effect, the confidence in the target increases exponentially.
Computational Approaches: Validating Targets at the Speed of Light
We are no longer limited by the physical speed of a pipette. In silico predictions and deep learning models can now screen millions of potential interactions in seconds. Pharmacophore models help us understand the 3D “map” of what a drug needs to look like to bind successfully, aiding in both target validation and drug repositioning. The advent of AlphaFold has further revolutionized this space by providing high-accuracy structural predictions for nearly every protein in the human proteome, allowing us to identify potential binding pockets that were previously invisible.
Network-based methods take this a step further. Instead of looking at one target in isolation, they map out the entire biological neighborhood—the interactome. This helps us predict if hitting one protein will cause a ripple effect of side effects elsewhere in the network. AI-powered molecular docking allows us to virtually “test-fit” drug molecules into target binding sites, providing quantitative scores for binding affinity before we ever enter the lab. This computational pre-screening ensures that only the most promising targets and compounds move into expensive wet-lab validation.
Why 75% of Published Targets Fail—And How to Spot Them
Here is a sobering fact: most of the “exciting” new targets published in top-tier journals cannot be reproduced in an industrial setting. The famous Bayer study, published in Nature Reviews Drug Discovery, revealed that only 20-25% of published target data held up under internal scrutiny. A similar study by Amgen found that only 6 out of 53 landmark oncology papers were reproducible. This “reproducibility crisis” is a major driver of clinical failure and wasted R&D budgets.
To combat this, modern validation programs use chemical proteomics and mass spectrometry to get an unbiased view of what a drug is actually doing. Instead of just looking at the target we hope it hits, these technologies look at every protein the drug touches. This is known as “target deconvolution.” It helps us spot off-target effects early and explains why some drugs work in mice but fail in humans due to subtle differences in protein structure or expression between species.
Interpretability and replicability challenges remain, but by integrating real-world evidence (RWE) and multi-omic data, we can build a much more reliable foundation. We must move away from relying on a single “breakthrough” paper and instead look for a convergence of evidence from multiple sources: genetic data, expression data, and functional assays. This “triangulation” of data is the hallmark of a sophisticated drug target validation strategy. It requires a culture of “healthy skepticism” where targets are aggressively challenged before they are allowed to progress into the lead optimization phase.
Beyond Averages: Use Single-Cell Data to Predict Clinical Success
The future of drug target validation is personal and granular. We are moving away from “bulk” analysis—which measures the average response of millions of cells—and toward single-cell analysis. This allows us to see how a target behaves in specific sub-populations of cells, such as rare cancer stem cells or specific neurons. This is critical for complex diseases like cancer or Alzheimer’s where cell heterogeneity is high; a drug might successfully hit a target in 90% of cells but fail because the 10% of cells it missed are the ones driving the disease.
Thermal proteome profiling (TPP) is another frontier. By combining the principles of CETSA with high-resolution mass spectrometry, we can monitor the thermal stability of the entire proteome at once. This tells us exactly which proteins a drug binds to, across thousands of candidates, in a single experiment. It provides a comprehensive map of both on-target and off-target interactions, allowing researchers to optimize compounds for maximum selectivity early in the process.
At Lifebit, we believe the key to the future lies in federated AI. The most valuable data for target validation is often locked away in secure silos—hospitals, biobanks, and research institutes. By enabling secure, real-time access to global biomedical and multi-omic data, we allow researchers to validate targets using diverse, real-world patient cohorts without moving sensitive data. This integration of AI-driven safety surveillance and patient-derived models ensures that the targets we choose today are the cures of tomorrow. We are moving toward a “digital twin” approach, where we can virtually simulate the effect of modulating a target in a specific patient’s biological network before a single dose is administered.
Solve Your Validation Hurdles: FAQ for Drug Discovery Teams
Why do most drugs fail in Phase II and III trials?
The most common reason is a lack of efficacy. This means the drug was safe enough to test in humans, but it didn’t actually treat the disease. This usually happens because the initial drug target validation was performed in models (like basic cell lines or certain animal models) that didn’t accurately represent human disease biology. If the target isn’t a primary driver of the disease in humans, even the most potent drug will fail to show clinical benefit.
How long does the target validation process typically take?
A standard drug target validation program typically takes between 2 and 6 months. This includes genetic studies, assay development, and initial pharmacological testing. While it might seem like a delay, it is a drop in the bucket compared to the 12-year drug development timeline. Investing this time upfront can save years of wasted effort and hundreds of millions of dollars on a non-viable target.
What is the difference between target identification and validation?
Target identification is about finding a “suspect”—a molecule that seems to be linked to a disease based on initial observations or data mining. Target validation is the “trial”—it’s the rigorous process of gathering enough evidence to prove that modulating that suspect will actually result in a therapeutic benefit for the patient. Identification is about correlation; validation is about causation.
What are “off-target” effects and how are they validated?
Off-target effects occur when a drug binds to proteins other than the intended target, potentially causing side effects. These are validated using techniques like chemical proteomics and broad-panel screening against known receptors and enzymes. Modern validation aims to identify these interactions as early as possible to guide the chemical optimization of the drug lead.
Can AI replace laboratory-based target validation?
AI is a powerful accelerator, but it cannot yet replace the lab entirely. AI can predict druggability, simulate binding, and analyze complex data sets to identify the best candidates for validation. However, the biological complexity of the human body still requires “wet-lab” confirmation in living cells or tissues to ensure that the AI’s predictions hold true in a real biological environment.
Stop Guessing: Secure Your Pipeline with Validated Targets
The math is simple: skipping or rushing drug target validation is the most expensive mistake a biotech or pharma company can make. In an era where Phase II attrition is 66%, we cannot afford to guess. We need robust, reproducible, and human-relevant data to guide our decisions. The industry is shifting toward a “fail fast, fail cheap” philosophy, where the goal is to identify non-viable targets as early as possible to preserve resources for the most promising breakthroughs.
Lifebit is here to transform how that data is accessed and analyzed. Our next-generation federated AI platform provides secure, real-time access to the global multi-omic data required for high-confidence validation. With our Trusted Research Environment (TRE) and Trusted Data Lakehouse (TDL), we enable secure collaboration across the globe, allowing you to harmonize data and run advanced AI/ML analytics where the data resides. This eliminates the need for risky and time-consuming data transfers, allowing your team to focus on the science.
By leveraging real-time insights and AI-driven safety surveillance through our R.E.A.L. layer, we help you identify the right targets faster and with more certainty. Don’t let your next breakthrough become another Phase II statistic. Let’s work together to build a future where every drug that enters the clinic has a validated path to success. By combining the best of human expertise with the power of federated AI, we can finally overcome the productivity crisis in drug discovery.
Ready to de-risk your discovery?

